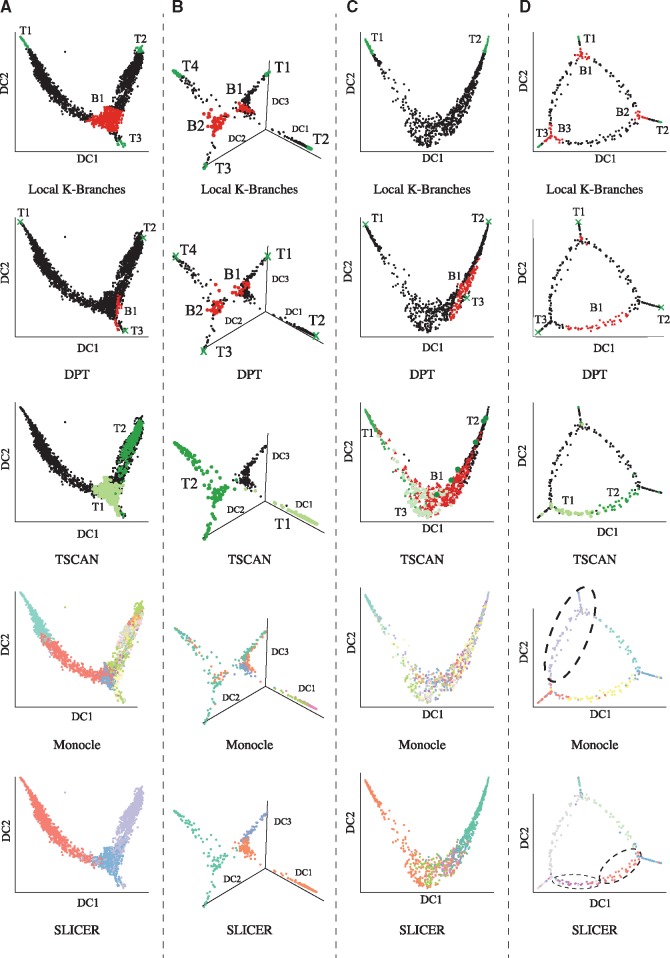
Fig. 4.
Results local K-Branches, DPT, TSCAN, Monocle and SLICER on all datasets. (A) Single-cell RNA-Seq data of myeloid progenitors (Paul et al., 2015). Common myeloid progenitor (CMP) cells branch into MEPs and GMPs. (B) Single-cell qPCR data of mouse blastocyst development (Guo et al., 2010), where the initial population of 2-cell blastocyst differentiates into three different groups of 64-cell blastocysts, undergoing two distinct branching events. (C) Single-cell qPCR data of human monocytic leukemia (Kouno et al., 2013). In this dataset, THP-1 human myeloid monocytic leukemia cells differentate into macrophages and no branching event is present. (D) Artificial data of arbitrary geometry where three distinct tips are connected by a loop