Fig. 1.
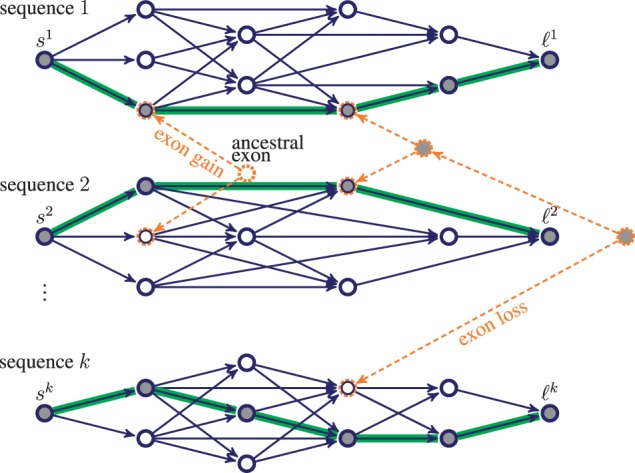
The joint gene structure graph G for a set of k homologous sequences. Nodes represent candidate exons. Blue/solid edges represent candidate introns or intergenic regions. Each path from the source si to the sink ℓi is a possible gene structure in sequence i. Homologous candidate exons are at the same time leaf nodes of phylogenetic trees (orange/dashed edges and nodes). A joint gene structure is sought: a binary labeling (filled, empty) of all nodes in G, whose restriction on the extant nodes (blue/solid) defines a collection of k paths (highlighted)
