Abstract
Insulin resistance (IR) is a precursor of type 2 diabetes (T2D), and improved risk prediction and understanding of the pathogenesis are needed. We used a novel high-throughput 92-protein assay to identify circulating biomarkers for HOMA of IR in two cohorts of community residents without diabetes (n = 1,367) (mean age 73 ± 3.6 years). Adjusted linear regression identified cathepsin D and confirmed six proteins (leptin, renin, interleukin-1 receptor antagonist [IL-1ra], hepatocyte growth factor, fatty acid–binding protein 4, and tissue plasminogen activator [t-PA]) as IR biomarkers. Mendelian randomization analysis indicated a positive causal effect of IR on t-PA concentrations. Two biomarkers, IL-1ra (hazard ratio [HR] 1.28, 95% CI 1.03–1.59) and t-PA (HR 1.30, 1.02–1.65) were associated with incident T2D, and t-PA predicted 5-year transition to hyperglycemia (odds ratio 1.30, 95% CI 1.02–1.65). Additional adjustment for fasting glucose rendered both coefficients insignificant and revealed an association between renin and T2D (HR 0.79, 0.62–0.99). LASSO regression suggested a risk model including IL-1ra, t-PA, and the Framingham Offspring Study T2D score, but prediction improvement was nonsignificant (difference in C-index 0.02, 95% CI −0.08 to 0.12) over the T2D score only. In conclusion, proteomic blood profiling indicated cathepsin D as a new IR biomarker and suggested a causal effect of IR on t-PA.
Introduction
Worldwide, diabetes affected over 387 million people and contributed to more than 4.9 million deaths in 2014. The prevalence of diabetes is projected to increase to 592 million by 2035 (1). Decreased sensitivity to circulating insulin (i.e., insulin resistance [IR]) induces compensatory hyperinsulinemia and leads to the development of type 2 diabetes (T2D) if pancreatic β-cell capacity is insufficient to maintain glucose homeostasis (2). IR constitutes both a precursor of and a therapeutic target in hyperglycemia and was found to be an independent risk factor for cardiovascular disease (CVD) (3), as well as a major contributor to vascular morbidity in T2D (4). Recent advances have made large-scale -omics studies possible that have pinpointed several tentative novel biomarkers for T2D, including branched-chain amino acids (5) and circulating microRNAs (6). Yet, a 2013 systematic review (7) failed to find evidence of benefit from adding novel circulating biomarkers and genetic markers to traditional T2D risk factors. Studies on biomarkers for IR have suggested several candidates, including ghrelin (8) and retinol-binding protein-4 (9). The identification of novel biologic predictors for T2D and IR is crucial for improved risk assessment and may help in understanding causal pathways beyond established genetic and lifestyle-related factors.
Recently, a new technology, the proximity extension assay (10), has enabled the simultaneous analysis of large sets of proteins in small biological sample volumes. We used such an immunoassay designed to analyze 92 proteins with proposed involvement in inflammation and CVD to explore potential biomarkers for IR. The objectives of this study were to 1) evaluate the association of CVD/inflammatory candidate protein biomarkers with prevalent IR in two large community cohorts without diabetes, 2) explore causal associations between biomarkers and IR in bidirectional Mendelian randomization (MR) analysis, and 3) assess the association of IR biomarkers with 10-year incident T2D and 5-year risk of transition to worse glycemia category, as well as to estimate the predictive performance of biomarkers for future T2D compared with an established risk score.
Research Design and Methods
Cohort Characteristics
Uppsala Longitudinal Study of Adult Men
In 1970, all male residents (n = 2,841) of Uppsala county, Sweden, born between 1920 and 1924 were invited to participate in the Uppsala Longitudinal Study of Adult Men (ULSAM) (n = 2,322 [81.7%] enrolled) (11), which includes regular assessments every 5–10 years. The baseline of the current study was set to the assessment at age 77 years (839 of 1,398 invited men [59.9%]), including recent targeted proteomic serum profiling. Diabetes was defined as fasting plasma glucose ≥7 mmol/L; HbA1c ≥6.5% (48 mmol/mol) at assessment ages 77, 82, and 88 years; use of antidiabetes medication according to the Swedish Prescribed Drug Register Anatomical Therapeutic Chemical classification code A10; or diagnosis of T2D according to the National Patient Register. Incident events of T2D were identified up to age 88 years. Proteomic profiling was done in 770 samples of which 8 were excluded during quality control. We excluded 156 individuals with prevalent diabetes and 66 with insufficient data for confounders, leaving 540 persons for inclusion in the current study. The regional ethics review board at Uppsala University approved the study, and all participants provided written informed consent.
Prospective Investigation of the Vasculature in Uppsala Seniors
In 2001, the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study group invited an unselected sample of 70-year-old residents of Uppsala community (1,016 of 2,025 invited persons [50.2%] were enrolled; 50% female) primarily to validate measures of endothelial function (12). Baseline assessment, including recent proteomic profiling of blood plasma, was done at age 70 years. Diabetes was defined as plasma glucose concentration ≥7 mmol/L at ages 70, 75, and 80 years; use of antidiabetes medication; or diagnosis of T2D according to validated hospital records. Incident T2D events were identified up to age 80 years. Among 1,003 subjects undergoing proteomic profiling, 12 were removed during quality control. We further excluded 116 individuals with prevalent diabetes and 48 with insufficient data for confounders, leaving 827 persons to be included in the current study. The regional ethics review board at Uppsala University approved the study, and all participants provided written informed consent.
Measurement of IR
The HOMA of IR index (HOMA-IR) was calculated according to the method proposed by Matthews et al. (13) with glucose in millimoles per liter and insulin in milliunits per liter (Eq. 1). Plasma insulin was measured by an ELISA in PIVUS (Boehringer, Mannheim, Germany) and ULSAM (Mercodia, Uppsala, Sweden). Glucose concentrations were quantified by the glucose dehydrogenase method (Gluc-DH by Merck, Darmstadt, Germany) in plasma from ULSAM and with similar methods (HemoCue, Ängelholm, Sweden) in whole blood from PIVUS (converted to plasma values by adding 11%).
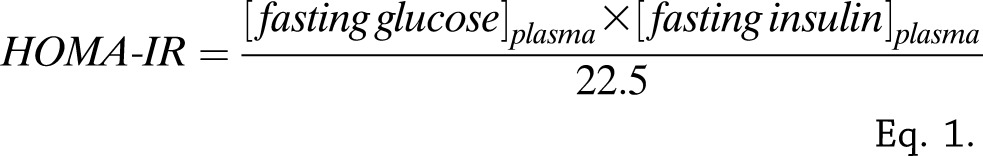 |
Proteomic Profiling
The Olink Proseek Multiplex CVD 96×96 proximity extension assay (10) uses two highly specific antibodies for each protein, which allows the formation of a PCR reporter sequence from attached oligonucleotide strands when both antibodies are bound to the target protein’s surface. The assay requires <10 μL sample volume and measures 92 proteins associated with CVD or inflammation and four internal control samples. Normalized protein expression (NPX) values were generated from quantitative PCR quantification cycle (Cq) values, where higher Cq corresponds to lower protein abundance. Cq values (log2 scale) were corrected for technical variation by an interplate control, and lower limits of detection (LOD) were determined through a negative control [NPX = Olink negative control − (ΔCqsample −Δinterplate control)]. The validation study of the assay, which included 90 proteins and seven samples analyzed in nine separate runs, found the mean intra-assay coefficient of variation to be 8% (range 4–13) and the interassay coefficient of variation to be 15% (range 11–39). Values below LOD were imputed as LOD/2, normalized for plate and storage time (based on the observed and predicted values obtained from a spline model) and rescaled to a distribution with a mean of 0 and an SD of 1. Quality control included removal of proteins with >15% samples below the LOD, and subjects with tail distribution (i.e., outlying) missingness as judged by histogram (>5% missing in PIVUS and >2% missing in ULSAM) were excluded. The final data set included 80 proteins. Proteins excluded from the statistical analyses were interleukin-4, melusin, natriuretic peptide B, β-nerve growth factor, SIR2-like protein, NF-κB essential modulator, pentraxin-related protein 3, N-terminal probrain natriuretic peptide, matrix metalloproteinase 7, membrane-bound aminopeptidase P, heat shock 27 kDa proteins, and cathepsin B.
Genetic Data
For MR analyses, we used the nonweighted genetic IR score composed of 10 single nucleotide polymorphisms (SNPs) validated in up to 18,565 subjects by Scott et al. (14) as an instrumental variable (IV) for HOMA-IR (Supplementary Table 1). We further identified suitable IVs for three biomarkers from a literature search (15–17). We then performed a genome-wide association study (GWAS) using the software SNPTEST v2.4 in the PIVUS and ULSAM studies for the remaining four biomarkers based on genotyping with the Illumina OmniExpress/Omni2.5 array combined with the Illumina Cardio-MetaboChip array, which was further imputed up to the 1000G March 2012 release using IMPUTE2 (18). The association of biomarker IVs with lnHOMA-IR was tested in the MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) cohort using publicly available data (19), and for the genetic IR score with biomarkers, we used PIVUS and ULSAM. For SNPs not reported in the MAGIC data, we selected a proxy variant in strong linkage disequilibrium (r2 > 0.8) via SNAP (http://www.broadinstitute.org/mpg/snap/ldsearch.php) and ascertained allele alignment with reference to the International HapMap Project CEU reference population (http://hapmap.ncbi.nlm.nih.gov).
Statistical Analysis
Association of Biomarkers with IR and Incident Diabetes Traits
All statistical analyses were carried out in R, version 3.1.1. Preliminary models indicated nonnormal distribution of model residuals, and C-reactive protein (CRP) concentrations and HOMA-IR were transformed to natural logarithmic scale to alleviate nonnormality. Separate linear regression models were assessed for each biomarker with lnHOMA-IR as the dependent variable and BMI, waist circumference, lnCRP, age, comorbidity, storage time, and sex as independent variables. Comorbidity was dummy coded based on a Charlson Comorbidity Index (20) of 0 or ≥1. The choice of the independent variables was based on a hypothetical causal diagram assisted by the DAGitty, version 2.2, software (www.dagitty.net) (21) (Supplementary Fig. 1). We first analyzed association in the PIVUS discovery sample, and those biomarkers passing the 5% false discovery rate (FDR) (22) were taken forward for replication in ULSAM. In the replication stage, 5% FDR was used again for determining significance. For all biomarkers, model assumptions of homoscedasticity and normality and the impact of potential outliers were examined in plots of residuals against normal quantiles (QQ-plot), fitted values, and leverage, respectively.
Thereafter, we assessed biomarkers related to IR for associations with 10-year incident T2D using Cox regression in ULSAM and PIVUS combined, with adjustment for BMI, waist, lnCRP, age, comorbidity, storage time, cohort, and sex. We additionally included fasting glucose levels in separate models. As the date of incident diabetes, the first recorded event fulfilling the definition of diabetes as specified above was used (i.e., date of diagnosis, antidiabetes medication prescription, or blood glucose thresholds at 5- and 10-year follow-ups). Individuals without an event were censored at the last assessment date or at date of death. The proportional hazards assumption was assessed using scaled Schoenfeld residual plots with formal significance testing for neutral slopes. Logistic regression analysis was used to predict 5-year risk of worsening fasting glycemia category (<5.6 mmol/L, 5.6–6.9 mmol/L, and ≥7 mmol/L or established T2D) in ULSAM from ages 77 to 82 years and in PIVUS from ages 70 to 75 years.
Predictive Performance and Comparison With Established Risk Factors
To assess predictive performance of biomarkers for 10-year T2D risk, we randomly split the combined cohorts of participants with sufficient data into a two-thirds learning (n = 911) and one-third internal validation sample (n = 456). The Framingham Offspring Study risk score for T2D (FORS, composed of sex, BMI, age, family history of diabetes, blood pressure, HDL cholesterol, triglycerides, and fasting glucose) (23) was calculated for each individual and used as a baseline model to assess the incremental improvement of adding biomarkers. Predictor selection was carried out in LASSO penalized Cox regression with 10-fold internal cross-validation in the learning sample and implemented with the glmnet package in R. In storage time– and cohort-adjusted models forced to include the FORS score, we allowed predictor choice among the validated IR biomarkers and used λ minimum to select the optimum model, which was then evaluated in the validation sample. We assessed discrimination (future case and noncase differentiation) with the receiver operating characteristic curve-based C-index (24) and compared models via likelihood ratio test. Calibration (the consistency between observed and predicted risks) was assessed by Grønnesby-Borgan test according to the methodology of May and Hosmer (25). This test is based on grouping subjects according to their risk estimates and comparing the sum of Cox model martingale residuals between groups, which assumes zero under the null hypothesis of perfect agreement between predicted and observed risks.
IV Analysis
Mendelian randomization techniques based on IV analysis were used to assess potential causal associations between biomarkers and IR in both directions (26).
Causal Effect of IR on Protein Concentrations
We evaluated the association of the genetic IR score with lnHOMA-IR using the summary statistics for 46,186 individuals without diabetes in the MAGIC cohort based on the method described by Dastani et al. (27) and implemented via gtx in R. The association of the genetic IR score with biomarkers was assessed in PIVUS and ULSAM separately in age- and sex-adjusted linear regression models and meta-analyzed using a fixed effect, SE-weighted model via metafor in R (Supplementary Table 2). The IV estimator βIV was calculated as the ratio of two regression coefficients based on the Wald ratio (βSNP-biomarker / βSNP–HOMA-IR). SEs were calculated using the Δ method, which we previously validated for use in a similar setting (28), as follows: abs (βIV) [(seSNP-intermediate / βSNP-intermediate)2 + (seSNP-outcome / βSNP-outcome)2]0.5. Causal estimators were tested at a nominal significance threshold of P < 0.05. Sensitivity analysis to exclude pleiotropy of the IV was performed by comparing IV estimates for individual SNPs in forest plots (Supplementary Fig. 2).
Causal Effect of Protein Concentrations on IR
We assessed the association of each biomarker IV with lnHOMA-IR in the MAGIC cohort (Supplementary Table 3). The association of each biomarker IV with biomarker concentration was derived from either published GWAS or ULSAM and PIVUS. The IV estimator βIV was calculated as βSNP–HOMA-IR / βSNP-biomarker, and SEs were calculated using the Δ method.
Results
Biomarkers for IR
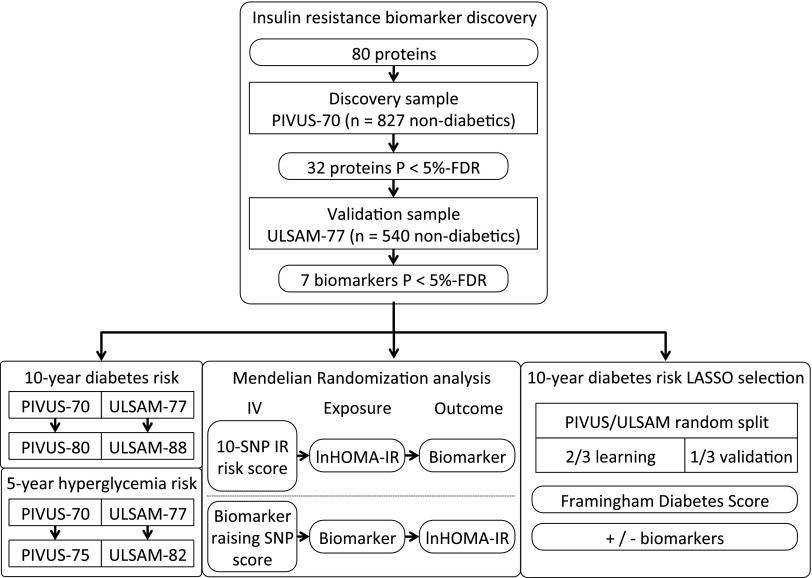
Table 1 shows baseline cohort characteristics. The design of the study is visualized in Fig. 1. We found 32 biomarkers associated with lnHOMA-IR in the PIVUS study (n = 827, 48.9% male, mean age 70.2 ± 0.2 years). Seven of these 32 biomarkers were replicated at a 5% FDR level in a sample of 540 men from the ULSAM cohort (mean age 77.6 ± 0.8 years). All subjects provided complete data for confounders. The seven identified biomarkers (leptin, tissue plasminogen activator [t-PA], renin, interleukin-1 receptor antagonist [IL-1ra], hepatocyte growth factor [HGF], cathepsin D, and fatty acid–binding protein 4 [FABP-4]) were all positively associated with IR (Table 2). Of these, leptin showed the strongest association (β 0.27, 95% CI 0.22–0.33 in PIVUS, and β 0.10, 95% CI 0.07–0.13 in ULSAM, where the coefficients represent the change in lnHOMA-IR associated with an SD-unit increase in NPX value).
Table 1.
Baseline cohort characteristics of participants without diabetes in PIVUS-70 and ULSAM-77
| PIVUS | N | ULSAM | N | |
|---|---|---|---|---|
| Women | 423 (51.1) | 827 | 0 | 540 |
| Age (years) | 70.2 ± 0.2 | 827 | 77.6 ± 0.8 | 540 |
| BMI (kg/m2) | 26.7 ± 4.1 | 827 | 26.0 ± 3.4 | 540 |
| CRP (mg/L) (ln transformed) | 0.62 ± 0.9 | 827 | 0.67 ± 1.0 | 540 |
| Waist circumference (cm) | 90.0 ± 11.0 | 827 | 94.6 ± 9.6 | 540 |
| Fasting glucose (mmol/L) | 5.5 ± 0.6 | 827 | 5.5 ± 0.6 | 540 |
| Fasting insulin (mU/L) | 8.3 ± 5.0 | 827 | 9.1 ± 8.0 | 540 |
| HOMA-IR | 2.1 ± 1.3 | 827 | 2.3 ± 2.1 | 540 |
| Systolic blood pressure (mmHg)* | 148.9 ± 22.5 | 823 | 150.3 ± 20.0 | 532 |
| Diastolic blood pressure (mmHg)* | 78.4 ± 10.0 | 823 | 81.1 ± 9.8 | 532 |
| Triglycerides (mmol/L) (ln transformed) | 0.1 ± 0.4 | 824 | 0.2 ± 0.4 | 539 |
| Subjects with comorbidities | 160 (19.3) | 827 | 208 (38.5) | 540 |
| Subjects with a 1st-degree relative with T2D | 115 (13.9) | 827 | 100 (18.5) | 540 |
| Length of follow-up (years) | 10.05 ± 0.17 | 827 | 9.13 ± 0.63 | 540 |
Data are mean ± SD or n (%) for categorical variables.
Assessed in either arm at rest using the routine sphygmomanometer technique.
Figure 1.
Flowchart illustrating the design of the study. P values were assessed at the 5% FDR. SNP genetic variants were used as IV.
Table 2.
Linear regression analysis results for biomarker associations with lnHOMA-IR, adjusted for age, sex, BMI, waist circumference, lnCRP, comorbidity, and storage time
| PIVUS (n = 827) | ULSAM (n = 540) | |||
|---|---|---|---|---|
| Biomarker | β (95% CI) | P | β (95% CI) | P |
| Leptin | 0.27 (0.22, 0.33) | 1.66 × 10−21 | 0.10 (0.07, 0.13) | 7.71 × 10−10 |
| t-PA | 0.11 (0.07, 0.14) | 5.97 × 10−9 | 0.06 (0.04, 0.09) | 2.54 × 10−7 |
| Renin | 0.12 (0.08, 0.15) | 4.22 × 10−11 | 0.05 (0.03, 0.07) | 6.30 × 10−5 |
| IL-1ra | 0.12 (0.08, 0.16) | 1.09 × 10−9 | 0.04 (0.02, 0.07) | 3.48 × 10−4 |
| HGF | 0.15 (0.12, 0.19) | 2.28 × 10−17 | 0.04 (0.02, 0.07) | 5.11 × 10−4 |
| Cathepsin D | 0.15 (0.11, 0.18) | 1.41 × 10−16 | 0.04 (0.02, 0.06) | 5.59 × 10−4 |
| FABP-4 | 0.16 (0.08, 0.17) | 2.20 × 10−8 | 0.04 (0.01, 0.06) | 7.67 × 10−3 |
β-Coefficients (95% CI) express the change in lnHOMA-IR associated with an SD-unit increase in NPX value. Raw P values are given for each association, and all proteins shown in this table are significant at the 5% FDR. Proteins with between 1 and 15% values below LOD in PIVUS were protein S100-A12 (13%), CD40 ligand (12%), TNF-related apoptosis-inducing ligand (9%), P-selectin glycoprotein ligand 1 (5%), caspase 8 (4%), leptin (4%), TNF-related activation-induced cytokine (3%), matrix metalloproteinase 3 (2%), pappalysin-1 (2%), FABP-4 (1%), and TNF ligand superfamily member 14 (1%); in ULSAM, the only protein was leptin (5%).
Bivariate Pearson correlations indicated positive associations (P < 0.05) between all seven biomarkers except for leptin and renin (r = 0.05, P = 0.058) in the low to moderate range (0.15–0.69).
Association With Incident Diabetes and Worsening Hyperglycemia
During follow-up (mean 9.7 ± 0.5 years), there were 73 and 38 incident cases of T2D in PIVUS and ULSAM, respectively, among 1,367 participants. In separate models adjusted for cohort and confounders, two biomarkers were associated with increased T2D risk (Table 3): IL-1ra (HR 1.28, 95% CI 1.03–1.59) and t-PA (HR 1.30, 95% CI 1.03–1.65). Additional adjustment for fasting glucose rendered both associations nonsignificant (t-PA HR 1.14, 95% CI 0.91–1.44; IL-1ra HR 1.19, 95% CI 0.94–1.50) and revealed a negative association with renin levels (HR 0.79, 95% CI 0.62–0.99).
Table 3.
Cox regression results for 10-year incident T2D and logistic regression results for 5-year risk of worse glycemia
| 10-year T2D risk (111 incident events) | 5-year worse glycemia risk (203 incident events) | |||
|---|---|---|---|---|
| Biomarker | HR (95% CI) | P | OR (95% CI) | P |
| Leptin | 1.39 (1.00, 1.95) | 0.054 | 1.02 (0.78, 1.33) | 0.909 |
| t-PA | 1.30 (1.03, 1.65) | 0.029 | 1.23 (1.02, 1.48) | 0.030 |
| Renin | 0.86 (0.68, 1.08) | 0.193 | 0.90 (0.75, 1.08) | 0.252 |
| IL-1ra | 1.28 (1.03, 1.59) | 0.025 | 1.04 (0.86, 1.25) | 0.681 |
| HGF | 1.21 (0.98, 1.51) | 0.082 | 0.98 (0.81, 1.18) | 0.828 |
| Cathepsin D | 1.23 (0.99, 1.53) | 0.058 | 0.99 (0.83, 1.19) | 0.936 |
| FABP-4 | 1.32 (0.99, 1.76) | 0.057 | 0.94 (0.75, 1.17) | 0.553 |
Adjusted HRs (adjusted for age, sex, BMI, waist circumference, lnCRP, storage time, and cohort) and odds ratios (OR) associated with an SD-unit increase in NPX value.
At the 5-year follow-up assessment, there were 115 and 88 cases of worse glycemic state compared with baseline in PIVUS and ULSAM, respectively. In adjusted logistic regression analysis, increased concentrations of t-PA (odds ratio 1.23, 95% CI 1.02–1.48) predicted worse glycemic status at 5-year follow-up (Table 3).
Comparison with Established Risk Factors for T2D
In LASSO Cox regression based on the learning sample, 10-year diabetes risk was predicted by a model that included the FORS score and the two biomarkers associated with T2D (t-PA and IL-1ra). In the internal validation set using the proposed β-coefficients, this new model improved the C-index compared with the FORS score–only model by 0.022, from C = 0.801 (95% CI 0.701–0.991) to C = 0.823 (95% CI 0.723–0.923). There was no significant difference in model fit (χ2 = 5.258, P = 0.07), with the biomarker model explaining 12.7% of the variance compared with 11.4% by the FORS score–only model. Both models demonstrated adequate calibration; Grønnesby-Borgan χ2 = 5.378, P = 0.25, for baseline and χ2 = 2.399, P = 0.66, for the full model.
Causal Associations Between IR and Biomarkers
In a literature search, we identified suitable IVs for IL-1ra (rs4251961 and rs6759676) (15), HGF (rs5745687) (16), and t-PA (rs9399599, rs3136739, and rs7301826) (17). In a GWAS for the remaining four biomarkers using the conventional threshold for genome-wide significance (P < 5 × 10−8), we found a suitable genetic IV for cathepsin D only (rs55861089) (Supplementary Table 3).
We found evidence of a causal effect of HOMA-IR on t-PA concentrations (βIV 3.21, 95% CI 0.72–5.70, P = 0.012). No evidence of a causal effect of IR on any of the other biomarkers was found (Supplementary Table 2). Sensitivity analysis for the genetic instrument did not indicate pleiotropic effects (Supplementary Fig. 2).
For the causal effect of biomarker on HOMA-IR, we identified suitable genetic IVs for IL-1ra, t-PA, HGF, and cathepsin D. The results of IV analysis in MAGIC did not show evidence of a causal effect of any of these biomarkers on IR (Supplementary Table 3).
Discussion
In two prospective community samples of 1,367 elderly individuals without diabetes, we identified seven proteins positively associated with prevalent IR, and one of these, cathepsin D, has not previously been reported as associated with IR. The correlations between the protein concentrations were weak to moderate. These correlations either could be explained by biomarkers being implicated in the same biological pathways or could represent different biological cascades related to IR and cardiovascular risk. We therefore carried all seven biomarkers forward for further analysis, where we found evidence for a causal effect of IR on t-PA concentrations suggesting an effect of IR on blood coagulation and extracellular matrix modeling—important components of atherosclerosis. We also found that higher baseline concentrations of t-PA and IL-1ra were associated with 10-year diabetes risk, and t-PA predicted worse 5-year fasting glucose levels. Compared with an established diabetes risk score, the addition of biomarkers did not improve discrimination significantly.
Causal Effect of IR on t-PA
MR analysis offers a statistical approach to inferring causality in observational studies. As variants of genetic alleles are randomly inherited at conception, their distributions are free from confounding influences and reverse causation. In MR analysis, a genetic variant or combination of variants known to be associated with an intermediate phenotype is used as the IV to assess the possible causal effect of the intermediate on the outcome variable (26).
Using MR analysis, we found evidence for a positive causal effect of IR on t-PA antigen levels, which has not previously been reported, although their correlation is well established (29). t-PA is expressed by endothelial cells and acts mainly by converting plasminogen to plasmin, thus contributing to fibrinolysis and extracellular matrix remodeling. It also acts as a proinflammatory cytokine (30). Circulating t-PA activity is regulated through complex formation with its main inhibitor, plasminogen activator inhibitor 1 (PAI-1), a major source of which is adipose tissue (31). Elevated PAI-1 activity and concentration are both associated with raised t-PA levels (32), and the observed causal effect of IR on t-PA antigen could be influenced by PAI-1 expression. Since we did not measure PAI-1 concentration or t-PA activity or differentiate between total and inhibitor-bound t-PA, we were not able to characterize the mechanisms in detail. Although recombinant t-PA is commonly used as a fibrinolytic drug, raised circulating t-PA levels are also a marker of future cardiovascular risk (33). The causal effect of IR on t-PA antigen suggested by our findings may contribute to the excess CVD risk in individuals with diabetes and requires validation in future studies.
Our study confirmed previous reports (32) of raised t-PA concentrations being associated with elevated T2D risk and extends these reports to an association with worsening 5-year fasting glycemia. Taken together, the current study confirms the role of t-PA in IR and T2D and points to a possible causal pathway from IR to t-PA concentrations.
A Novel Association Between Cathepsin D and IR
For six of the identified protein markers, we confirm previously reported associations with IR in humans (29,34–38). However, the seventh protein, cathepsin D, has to our knowledge not previously been linked to IR.
The lysosomal endopeptidase cathepsin D is expressed ubiquitously, and its main effects include intracellular protein turnover and extracellular matrix breakdown. Altered expression of the protein has been implicated in, for example, Alzheimer disease, atherosclerosis, and breast cancer (39).
Raised free fatty acid levels and advanced glycation end products found in prediabetes states have recently been shown to enhance cathepsin D release (40–43). This may contribute to IR through mitochondrial dysfunction (42), impaired detoxification of advanced glycation end products (43), and the induction of proapoptotic proteins (44).
As weight gain was shown to stimulate its activity leading to adipocyte apoptosis, cathepsin D was suggested as a potential mediator between obesity and chronic adipose tissue inflammation (44), an important contributor to IR (2). The observed strong association between cathepsin D and IR in the current study may be the result of the deranged intracellular homeostasis resulting from lipotoxicity and inflammation in insulin-resistant states. Our MR study did not support a causal effect in any of the two directions, and we cannot exclude that the association we identified could be due to unmeasured confounding.
Cathepsin D as a possible mediator between overweight, inflammation, and metabolic disease may be amenable to drug targeting, and recent advances have been made in the field of cancer (45). Although we found a strong link between cathepsin D and prevalent IR, we failed to detect an association with 10-year diabetes risk or with 5-year worsening hyperglycemia. Future studies on the implication of cathepsin D in diabetes are needed.
Protein Biomarkers for Future Diabetes and Hyperglycemia
We confirmed the previously reported association of IL-1ra concentrations with T2D risk. IL-1ra competitively inhibits IL-1 from binding to its receptor, thereby suppressing its proinflammatory effects. In a retrospective analysis unadjusted for baseline glucose levels, IL-1ra was elevated up to 13 years prior to T2D diagnosis (46), which is in agreement with the association with 10-year incidence of T2D. Adjustment for fasting glucose resulted in a positive but insignificant association in the current study. Carstensen et al. (46) argued against adjusting for fasting glucose on the basis of it forming part of the definition of incident T2D. Although initially, recombinant IL-1ra agonists improved glycemic and inflammatory measures in T2D patients (47), long-term benefits have yet to be demonstrated, and concerns about cardiovascular side effects from increased IL-1ra levels have been raised (48).
Clinical Implications
The addition of IR biomarkers to the FORS score did not improve prediction. However, the number of events in our study was moderate, and larger studies are needed for more precise estimates. The observed associations with IR support the prospective validation of the assay for translating targeted proteomics into the diabetes care practice, but no direct clinical implications in the short term should arise from our findings. However, the identification of cathepsin D as an IR risk protein in this proteomics study in large community samples suggests potential benefits of applying this technology to biomarker discovery in the clinical setting and for other pathologies.
Limitations
Both cohorts are demographically homogeneous and consist of elderly persons, thus limiting generalizability to other ethnic and age groups. A fasting blood sample–based proxy measure (HOMA-IR) was used in this study, which does not provide a perfect reflection of the physiology of IR. The scale of the proteomics assay is not readily convertible to absolute concentrations for comparisons with previous studies. While we attempted to reduce bias in MR modeling by, e.g., sensitivity analysis for pleiotropy of the genetic IR score, our findings are limited by the lack of statistical power for the IR-biomarker section. Since for cathepsin D and HGF, the genetic variant used as IV maps to the biomarker’s coding region, possible false signals due to interference with assay antibody binding that could have resulted in an invalid instrument cannot be excluded. Finally, the assay used focused on proteins associated with CVD and/or inflammation and was not specifically targeted toward metabolism. An assay targeted directly toward diabetes candidate proteins may have revealed additional findings.
Conclusion
We found evidence of a causal effect of IR on t-PA antigen concentrations, which could be part of the explanation of the excess risk of CVD in the population with diabetes. We further identified cathepsin D as a novel potential biomarker for IR and demonstrated the application of high-efficiency targeted proteomics for diabetes risk assessment.
Supplementary Material
Article Information
Acknowledgments. The authors thank Liisa Byberg, Uppsala University, for assistance with calculation of the comorbidity score in ULSAM. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, Sweden.
Funding. This study was supported by the Swedish Research Council (Vetenskapsrådet, grant no. 2012-1397), the Swedish Heart-Lung Foundation (20140422), the Swedish Diabetes Foundation (Diabetesfonden) (grant no. 2013-024), the Knut och Alice Wallenberg Foundation, the European Research Council (ERC Starting Grant), and Uppsala University.
Duality of Interest. T.F. has received honoraria for lecturing from Merck Sharp & Dohme. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. C.N. implemented and planned the statistical analysis and wrote the manuscript. J.S., S.G., E.I., and T.F. planned the statistical analysis. J.S., S.G., L.L., E.I., and T.F. revised the manuscript. J.S., V.G., L.L., and E.I. participated in data acquisition. E.I. and T.F. conceived the study. T.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 75th Scientific Sessions of the American Diabetes Association, Boston, MA, 5–9 June 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0881/-/DC1.
References
- 1.International Diabetes Federation IDF Diabetes Atlas. 6th ed. Brussels, Belgium, International Diabetes Federation, 2014, p. 32–39 [Google Scholar]
- 2.Dali-Youcef N, Mecili M, Ricci R, Andrès E. Metabolic inflammation: connecting obesity and insulin resistance. Ann Med 2013;45:242–253 [DOI] [PubMed] [Google Scholar]
- 3.Bonora E, Formentini G, Calcaterra F, et al. . HOMA-estimated insulin resistance is an independent predictor of cardiovascular disease in type 2 diabetic subjects: prospective data from the Verona Diabetes Complications Study. Diabetes Care 2002;25:1135–1141 [DOI] [PubMed] [Google Scholar]
- 4.Park SW, Kim SK, Cho YW, et al. . Insulin resistance and carotid atherosclerosis in patients with type 2 diabetes. Atherosclerosis 2009;205:309–313 [DOI] [PubMed] [Google Scholar]
- 5.Batch BC, Hyland K, Svetkey LP. Branch chain amino acids: biomarkers of health and disease. Curr Opin Clin Nutr Metab Care 2014;17:86–89 [DOI] [PubMed] [Google Scholar]
- 6.Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 2013;9:513–521 [DOI] [PubMed] [Google Scholar]
- 7.Echouffo-Tcheugui JB, Dieffenbach SD, Kengne AP. Added value of novel circulating and genetic biomarkers in type 2 diabetes prediction: a systematic review. Diabetes Res Clin Pract 2013;101:255–269 [DOI] [PubMed] [Google Scholar]
- 8.Barazzoni R, Zanetti M, Ferreira C, et al. . Relationships between desacylated and acylated ghrelin and insulin sensitivity in the metabolic syndrome. J Clin Endocrinol Metab 2007;92:3935–3940 [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Graham TE, Mody N, et al. . Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356–362 [DOI] [PubMed] [Google Scholar]
- 10.Assarsson E, Lundberg M, Holmquist G, et al. . Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedstrand H. A study of middle-aged men with particular reference to risk factors for cardiovascular disease. Ups J Med Sci Suppl 1975;19:1–61 [PubMed] [Google Scholar]
- 12.Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to determine arterial compliance in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. J Hypertens 2006;24:1075–1082 [DOI] [PubMed] [Google Scholar]
- 13.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 14.Scott RA, Fall T, Pasko D, et al.; RISC Study Group; EPIC-InterAct Consortium . Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes 2014;63:4378–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herder C, Nuotio ML, Shah S, et al. . Genetic determinants of circulating interleukin-1 receptor antagonist levels and their association with glycemic traits. Diabetes 2014;63:4343–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieb W, Chen MH, Larson MG, et al. . Genome-wide association study for endothelial growth factors. Circ Cardiovasc Genet 2015;8:389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Huffman JE, Yamakuchi M, et al.; Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium Neurology Working Group; CARDIoGRAM Consortium; CHARGE Consortium Hemostatic Factor Working Group . Genome-wide association study for circulating tissue plasminogen activator levels and functional follow-up implicates endothelial STXBP5 and STX2. Arterioscler Thromb Vasc Biol 2014;34:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parihar A, Wood GC, Chu X, et al. . Extension of GWAS results for lipid-related phenotypes to extreme obesity using electronic health record (EHR) data and the Metabochip. Front Genet 2014;5:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987;40:373–383 [DOI] [PubMed] [Google Scholar]
- 21.Pearl J. Causality: Model, Reasoning, and Inference. Cambridge, U.K., Cambridge University Press, 2009, p. 41–64 [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300 [Google Scholar]
- 23.Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 2007;167:1068–1074 [DOI] [PubMed] [Google Scholar]
- 24.Harrell FE Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA 1982;247:2543–2546 [PubMed] [Google Scholar]
- 25.May S, Hosmer DW. A simplified method of calculating an overall goodness-of-fit test for the Cox proportional hazards model. Lifetime Data Anal 1998;4:109–120 [DOI] [PubMed] [Google Scholar]
- 26.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res 2007;16:309–330 [DOI] [PubMed] [Google Scholar]
- 27.Dastani Z, Hivert MF, Timpson N, et al.; DIAGRAM+ Consortium; MAGIC Consortium; GLGC Investigators; MuTHER Consortium; DIAGRAM Consortium; GIANT Consortium; Global B Pgen Consortium; Procardis Consortium; MAGIC investigators; GLGC Consortium . Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet 2012;8:e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fall T, Hägg S, Mägi R, et al.; European Network for Genetic and Genomic Epidemiology (ENGAGE) consortium . The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med 2013;10:e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amowitz LL, Manson JE, Ridker PM. Association of endogenous tissue plasminogen activator (t-PA) with clinical characteristics of the insulin resistance syndrome. J Thromb Thrombolysis 2000;10:227–231 [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Hu K. Tissue plasminogen activator and inflammation: from phenotype to signaling mechanisms. Am J Clin Exp Immunol 2014;3:30–36 [PMC free article] [PubMed] [Google Scholar]
- 31.Lundgren CH, Brown SL, Nordt TK, Sobel BE, Fujii S. Elaboration of type-1 plasminogen activator inhibitor from adipocytes. A potential pathogenetic link between obesity and cardiovascular disease. Circulation 1996;93:106–110 [DOI] [PubMed] [Google Scholar]
- 32.Eliasson MC, Jansson JH, Lindahl B, Stegmayr B. High levels of tissue plasminogen activator (tPA) antigen precede the development of type 2 diabetes in a longitudinal population study. The Northern Sweden MONICA study. Cardiovasc Diabetol 2003;2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freynhofer MK, Draxler DF, Gruber SC, et al. . Endogenous t-PA-antigen is an independent predictor of adverse cardiovascular events and all-cause death in patients with atrial fibrillation. J Thromb Haemost 2013;11:1069–1077 [DOI] [PubMed] [Google Scholar]
- 34.Zuo H, Shi Z, Yuan B, Dai Y, Wu G, Hussain A. Association between serum leptin concentrations and insulin resistance: a population-based study from China. PLoS One 2013;8:e54615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lind L, Reneland R, Andersson PE, Haenni A, Lithell H. Insulin resistance in essential hypertension is related to plasma renin activity. J Hum Hypertens 1998;12:379–382 [DOI] [PubMed] [Google Scholar]
- 36.Fuseya T, Furuhashi M, Yuda S, et al. . Elevation of circulating fatty acid-binding protein 4 is independently associated with left ventricular diastolic dysfunction in a general population. Cardiovasc Diabetol 2014;13:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukagawa E, Adachi H, Hirai Y, et al. . Independent association of elevated serum hepatocyte growth factor levels with development of insulin resistance in a 10-year prospective study. Clin Endocrinol (Oxf) 2013;79:43–48 [DOI] [PubMed] [Google Scholar]
- 38.Charles BA, Doumatey A, Huang H, et al. . The roles of IL-6, IL-10, and IL-1RA in obesity and insulin resistance in African-Americans. J Clin Endocrinol Metab 2011;96:E2018–E2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masson O, Bach AS, Derocq D, et al. . Pathophysiological functions of cathepsin D: Targeting its catalytic activity versus its protein binding activity? Biochimie 2010;92:1635–1643 [DOI] [PubMed] [Google Scholar]
- 40.Tan KC, Shiu SW, Wong Y, Tam X. Serum advanced glycation end products (AGEs) are associated with insulin resistance. Diabetes Metab Res Rev 2011;27:488–492 [DOI] [PubMed] [Google Scholar]
- 41.Almaguel FG, Liu JW, Pacheco FJ, De Leon D, Casiano CA, De Leon M. Lipotoxicity-mediated cell dysfunction and death involve lysosomal membrane permeabilization and cathepsin L activity. Brain Res 2010;1318:133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boya P, Andreau K, Poncet D, et al. . Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J Exp Med 2003;197:1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm S, Horlacher M, Catalgol B, Hoehn A, Reinheckel T, Grune T. Cathepsins D and L reduce the toxicity of advanced glycation end products. Free Radic Biol Med 2012;52:1011–1023 [DOI] [PubMed] [Google Scholar]
- 44.Eguchi A, Feldstein AE. Lysosomal Cathepsin D contributes to cell death during adipocyte hypertrophy. Adipocyte 2013;2:170–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maynadier M, Vezenkov LL, Amblard M, et al. . Dipeptide mimic oligomer transporter mediates intracellular delivery of Cathepsin D inhibitors: a potential target for cancer therapy. J Control Release 2013;171:251–257 [DOI] [PubMed] [Google Scholar]
- 46.Carstensen M, Herder C, Kivimäki M, et al. . Accelerated increase in serum interleukin-1 receptor antagonist starts 6 years before diagnosis of type 2 diabetes: Whitehall II prospective cohort study. Diabetes 2010;59:1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen CM, Faulenbach M, Vaag A, Ehses JA, Donath MY, Mandrup-Poulsen T. Sustained effects of interleukin-1 receptor antagonist treatment in type 2 diabetes. Diabetes Care 2009;32:1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Interleukin 1 Genetics Consortium Cardiometabolic effects of genetic upregulation of the interleukin 1 receptor antagonist: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol 2015;3:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.