Fig. 1.
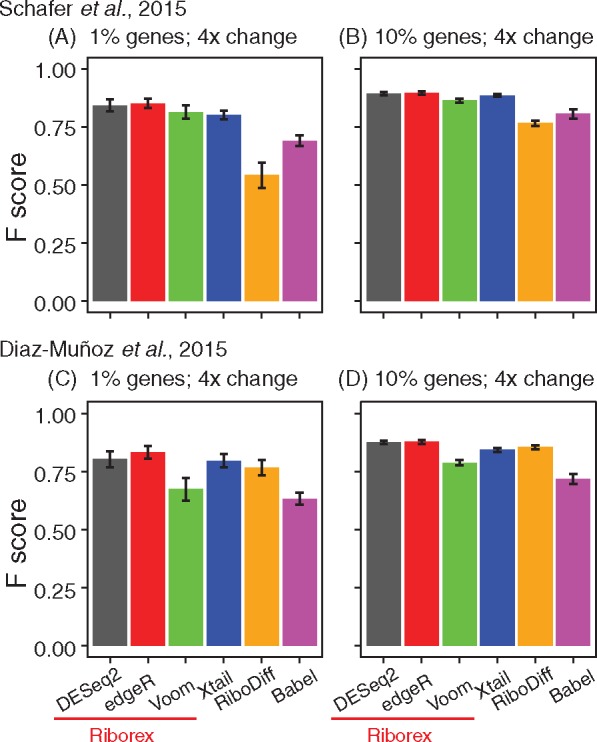
Accuracy identifying differentially translated genes in simulated data (see supp. info.) comparing Riborex (using DESeq2, edgeR and Voom (Law et al., 2014)), Xtail, RiboDiff and Babel. Values are averages of 100 simulations and error bars indicate standard deviation of F scores. (A) and (B) Results based on rat heart data (Schafer et al., 2015) with 1% and 10% implanted true differentially translated genes, respectively, and simulated 4-fold change in translation efficiency. (C) and (D) Results from simulations based on mouse HuR data (Diaz-Muñoz et al., 2015)
