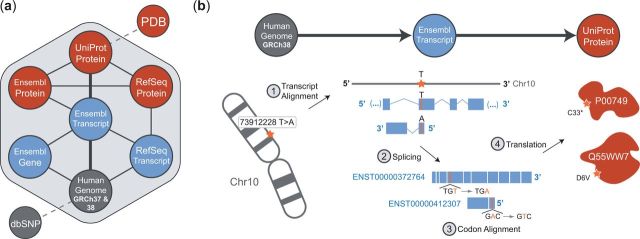
Fig. 1.
(a) The core BISQUE conversion graph, including all possible starting and ending points for a conversion with BISQUE. (b) The steps for conversion of a genomic variant to UniProt amino acid substitution(s) by traversing a conversion path through Ensembl transcript. The shown genomic variant maps to two Ensembl transcripts, one on the forward strand and one on the reverse. BISQUE uses genomic alignments of transcripts from Ensembl’s database, removes introns and non-coding exons (UTRs and alternatively spliced regions) and codon-aligns the result to match amino acids in UniProt proteins