Fig. 1.
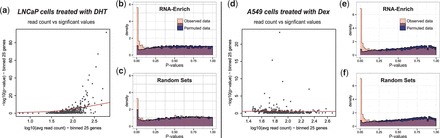
(a) RNA-seq data from LNCaP cells treated with DHT compared with a control showed a relationship between average gene read count and −log10(P-values) from DE tests. (b–c) Histogram of P-values from the permuted data should be uniformly distributed for acceptable type I error rate. For RNA-Enrich, the type I error rate is approximately uniform (b), but for the random sets approach for which there is no correction, more P-values are significant than expected (c). With the original data, RNA-Enrich identifies more significant GO terms than the random sets method in the observed data. (d) RNA-seq data from A549 cells treated with Dex compared with ethanol showed no relationship between read count and −log10(P-values). (e–f) With or without the read count bias correction, type I error rate is approximately uniform, indicating that no correction is needed and either test is valid. Histograms are transparent and overlaid. Additional permutations, enrichment testing methods and dataset are provided in the supplement
