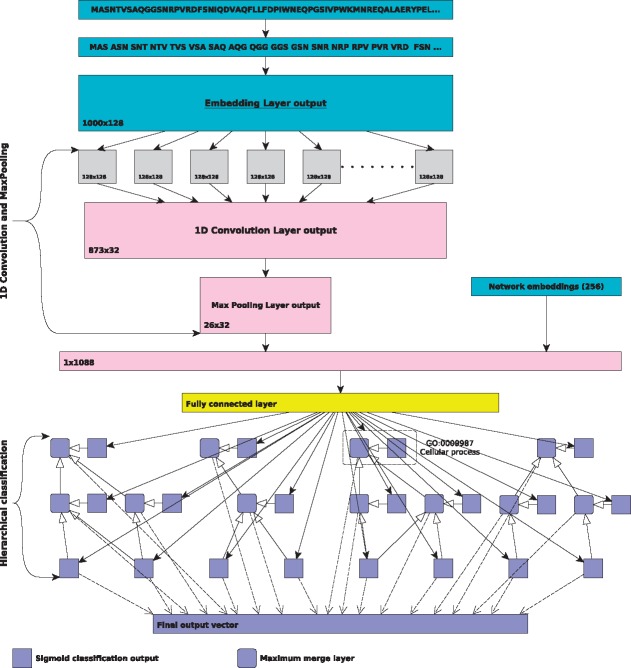
Fig. 1.
Convolutional Neural Network Architecture. (1) The input of the model is a list of integer indexes of trig.rams generated from protein sequence and vector of size 256 for protein PPI network representation. The trigram indexes are passed to an embedding layer which provides vector representations of size 128 for each trigram. The output of an embedding layer is a matrix of size 1000 × 128 on which we apply convolution and max-pooling. We merge the flattened output of the max-pooling layer and concatenate the resulting vector with the PPI network embeddings. This feature vector is then passed to hierarchically structured classification layers. (2) The hierarchically structured classification layers form a directed acyclic graph following the taxonomic structure of GO for is-a relations. For each GO class we generate one fully connected layer with a sigmoid activation function that predicts whether the input should be classified with this GO class. To ensure consistency, all non-leaf nodes in the graph use a maximum merge layers (rounded purple square) which outputs the maximum value of the classification results for all child nodes and the internal node’s classification results. The output vector of the model is the concatenation of maximum merge layers of the internal nodes and the classification layers of the leaf nodes