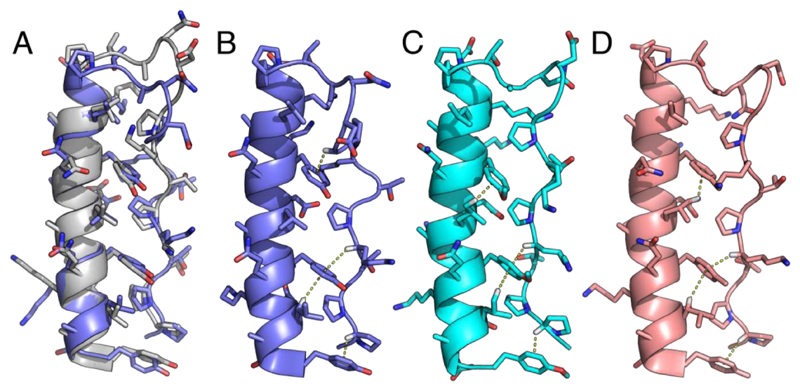
Figure 3. NMR structures for the p-substituted phenylalanine variants of PPα.
A: NMR structure closest to the geometric mean of the ensemble (model 20) for PPα-Tyr (slate) overlaid with the in silico model after 100 ns of MD (gray). B-D: Representative NMR structures from the ensembles showing the CH–π interactions found for PPα-Tyr, model 14 (B), PPα-ϕOCH3, model 8 (C), and PPα-ϕCH3, model 5 (D). The average numbers of CH–π interactions per ensemble structure were 2.25, 2.7 and 2.5, respectively, with 1.2, 0.65 and 0.55 per structure involving Pro. Although the remaining PPα peptides were folded by NMR they gave poor-quality spectra and structure calculations were not possible, which corroborated their reduced thermal stability. PDB codes: PPα-Tyr, 5LO2; PPα-ϕOCH3, 5LO3; and PPα-ϕCH3, 5LO4.