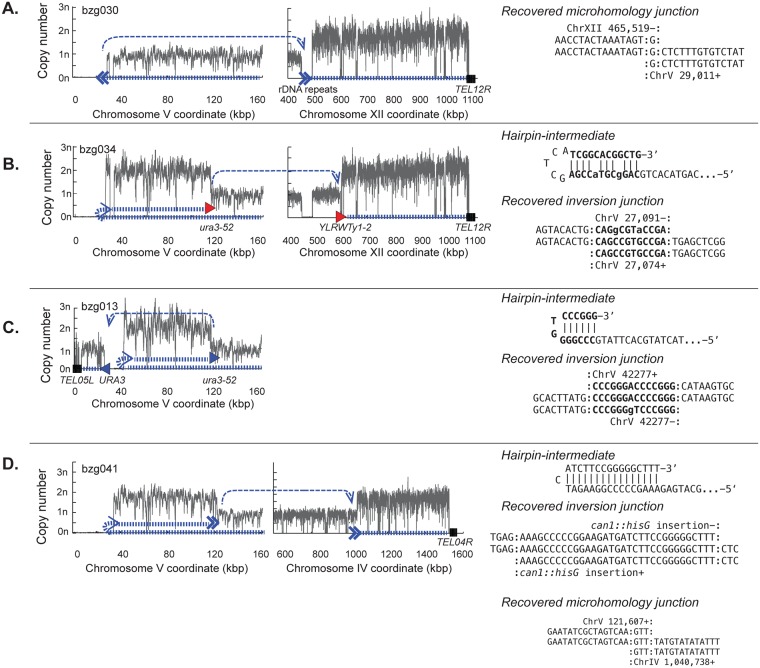
Fig 3. Types of GCRs identified by whole genome sequencing.
Analysis of a microhomology-mediated translocation (A) and hairpin-mediated inverted duplications that were resolved by homologous recombination between Ty-derived elements (B), the homology between URA3 and ura3-52 (C), and a microhomology-mediated translocation (D). Left. Copy number analysis of uniquely mapping regions along a portion of the left arm of the assay-containing chromosome V derived from the read depth of uniquely mapping regions of the genome. Middle. Copy number analysis that, if present, shows other copy number changes elsewhere in the genome. Left and Middle. The path describing the GCR-containing chromosome is illustrated by the hashed thick blue line; the thin dashed blue lines indicate the connectivity between individual fragments that are separated on the reference genome. Homology-mediated translocation junctions are depicted with filled in triangles that point in the direction in which homology element points; junctions involving Ty-related homologies are red and other homologies are blue. Non-homology or micro-homology translocations are shown using two chevrons. Telomeres associated with the GCR (if known) are shown as a series of black vertical lines. Right. Sequences of any novel junctions are with the central line in the alignment corresponding to the novel junction. Sequences at the junction that could have been derived from either sequence are surrounded with colons. For GCRs with hairpin-mediated inversions, the inferred structure of the hairpin intermediate is also shown.