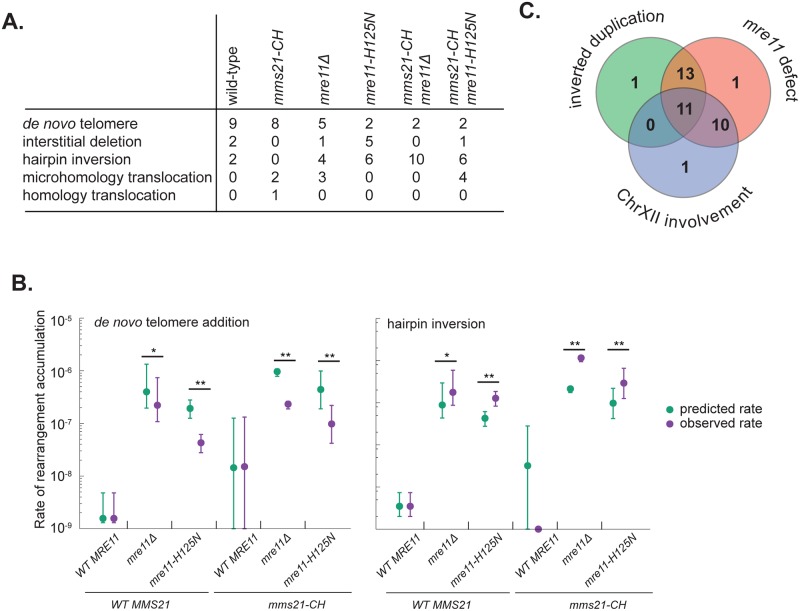
Fig 4. Distribution of GCR types from each mutant analyzed.
A. Table of the number of each type of rearrangement, de novo telomere addition, interstitial deletion, hairpin-mediated inverted duplication, micro- and non-homology-mediated translocations, and homology-mediated translocations. B. The observed rates (purple) of accumulating de novo telomere addition GCRs and hairpin inversion GCRs were calculated by multiplying the observed rate and 95% confidence intervals by the fraction of GCRs in each mutant corresponding to each type of GCR. The expected rates (green) were calculated by multiplying the rates by the fraction of GCRs in the wild-type strain. Statistically significant differences between the observed and expected rates were calculated by the Mann-Whitney test; p-values < 0.05 displayed as ‘*’ and p-values < 0.0005 displayed as ‘**’. C. Venn diagram of sequenced GCRs demonstrates that strains with MRE11 defects tend to accumulate GCRs involving inverted duplications and/or chromosome XII.