Fig. 1. Expanding the repertoire of ordered macrocycles by comprehensive sampling.
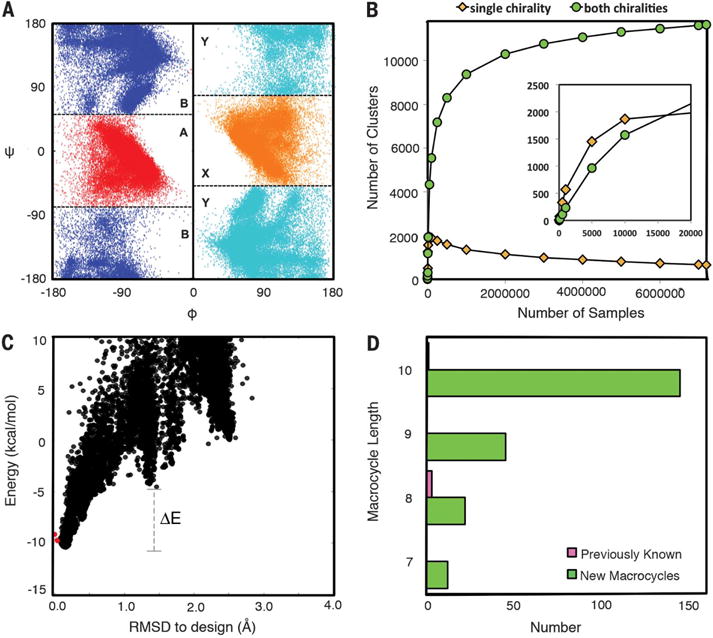
(A) Torsion bin definitions (bin B = blue; A = red; Y = mirror of bin B, cyan; X = mirror of bin A, orange) overlaid on the generated nine-residue macrocycle backbone torsion angle distributions. The inversion symmetry is evident. (B) Convergence test. The number of clusters (torsion bin strings) observed for which the mirror image is not observed (yellow) or for which the mirror image is observed (cyan) was determined both for the complete set of 7,500,000 samples for nine-residue macrocycles and for randomly selected subsets of different sizes (x axis). For comparison, the less converged results obtained starting from a 20,000-sample subset and then down-sampling are shown in the inset: The fraction of single-chirality clusters is higher, and the number of both-chirality clusters does not plateau. (C) Energy landscape analysis using large-scale genKIC backbone generation followed by all-atom energy minimization. Each point is the result of an independent calculation; the energy is shown on the y axis and the root mean square deviation (RMSD) from the design model on the x axis. The extent of funneling (or convergence) of the energy landscape is quantified through the energy gap between conformations generated by stochastic sampling around the design model (red points) and high-RMSD structures, and the Boltzmann weight of the near–design model population. (D) Number of designed macrocycle clusters with distinct torsion bin strings and backbone hydrogen bond patterns with energy gaps less than −0.1 and Boltzmann weights greater than 0.85; the number of unbound peptide macrocycles in the PDB and CSD, consisting of only 20 canonical amino acids and their D-enantiomers, is indicated in pink.
