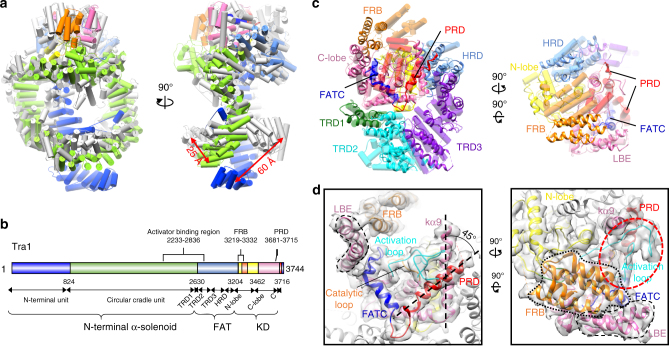
Fig. 3.
Structure of the Tra1 in the TEEAA assembly. a Structure comparison of Tra1 with DNA-PKcs. The Tra1 subunit is color-coded by domain assignment as panel b and aligned with the crystal structure of DNA-PKcs (gray, PDB ID: 5LUQ). Significant differences in the two structures are observed in the N-terminal HEAT repeats. The N-terminal unit of Tra1 moves inwards by 60 Å, while the circular cradle unit moves a smaller distance by 25 Å. b A schematic representation highlighting the functional domains of Tra1. Three units of Tra1: N-terminal HEAT repeat (the N-terminal unit and the circular cradle unit), the FAT (TRD1, 2, 3 and HRD), and the kinase regions are labeled. The activator binding region, the FRB insertion and the PRD domain are labeled above the schematic. c Structural comparison of the FATKIN region of Tra1 and DNA-PKcs. Tra1 FATKIN is shown in ribbon diagram model and DNA-PKcs FATKIN is shown in transparent pipes. The domain organization and color assignment are the same as Supplementary Fig. 7. d Two detailed views of the active site of Tra1, with the N-lobe is shown in yellow, the C-lobe in hot pink, the FRB insertion in orange, PRD in red, and FATC in blue. The cryo-EM density is displayed in transparent surface. The activation loop is colored in cyan and the catalytic loop in orange. The FAT domain is shown in gray. The LBE domain is highlighted in black dashed lines, while the FRB insertion in dotted lines. PRD moves outwards by 45° relative to the kα9. The red dashed circle highlights the highly extended and exposed activation loop