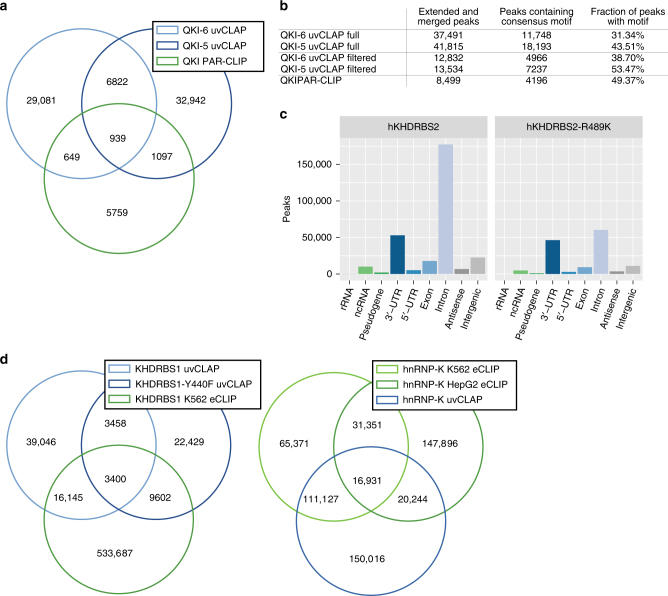
Fig. 5.
Comparison of uvCLAP to other methods. a Overlap between the full sets of QKI-5 and QKI-6 uvCLAP and QKI PAR-CLIP peaks. Peaks were extended to a minimum length of 41 nt, adjacent or overlapping sites were merged. b QKI-5, QKI-6 and QKI PAR-CLIP sites containing the consensus motif AYUAAY. c Number of JAMM peaks located on genomic target classes. Each peak was assigned once to the category with highest priority corresponding to the order rRNA, ncRNA, pseudogene, 3’-UTR, 5’-UTR, exon, intron, antisense, intergenic. d (left) Overlap between the full sets of KHDRBS1, KHDRBS1Y440F uvCLAP and KHDRBS1 K563 eCLIP peaks. (right) Overlap between the full sets of hnRNPK uvCLAP, hnRNPK K562 eCLIP and hnRNPK HepG2 eCLIP peaks. Peaks were extended to a minimum length of 41 nt, overlapping or abutting sites were merged