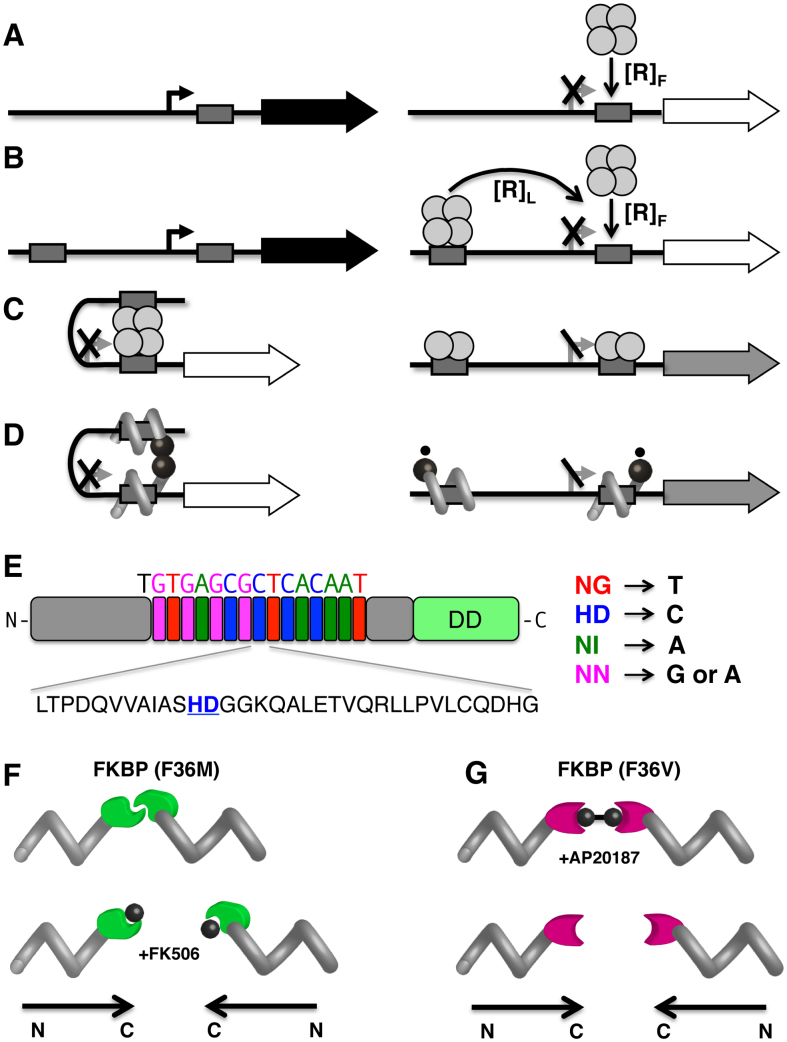
Figure 1.
Engineering DNA regulatory loops by analogy with natural bacterial repressors. (A) Constitutive ‘off’ switch. ‘On’ state (left) involves an unoccupied operator (grey box) at the promoter (broken arrow) allowing transcription of the downstream gene (filled arrow). ‘Off" state (right) involves a repressor protein (shown as a tetramer) binding at the operator, preventing promoter access by RNA polymerase. (B) Enhancing effective repressor concentration by DNA looping. Two unoccupied operators (grey boxes) flank the promoter to be regulated (left). The effective concentration of repressor (shown as a bidentate tetramer) at the proximal regulatory operator is the sum of contributions due to free, [R]F, and tethered, [R]L (distal) repressors (right). (C) Inducible ‘off’ switch. ‘Off’ state (left) involves DNA looping anchored by a bidentate tetrameric repressor protein. ‘On’ state (right) is induced by tetramer destabilization into dimers, removing the contribution of tethered (distal) repressors. Residual repression is indicated by grey arrow. (D) Analogous inducible ‘off’ switch involving a designed TALED. A small molecule (small circles) decreases dimer stability. (E) TALED design. TALE protein (gray) with sequence-specific RVD cassettes highlighted and the amino acid sequence of one cassette illustrated (below) with specificity diresidue underlined. DD: dimerization domain. (F) Controlled TALED dimerization. TALE-FKBP(F36M) dimerization variant (left) is broken by FK506 (small circle). Polypeptide polarity is shown below. (G) TALE-FKBP(F36V) dimerization variant (right) is induced by AP20187.