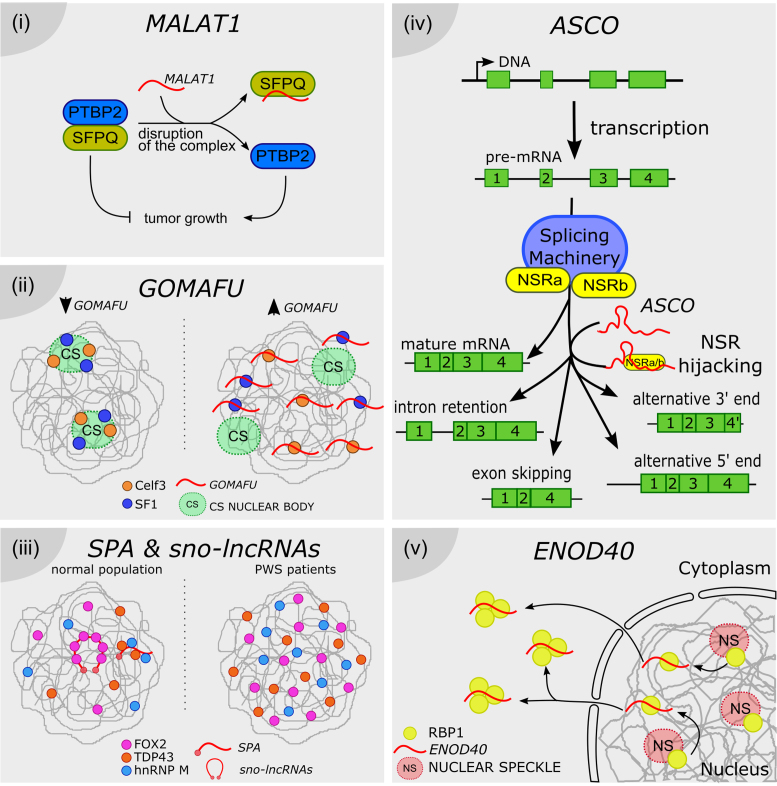
Figure 2.
Long noncoding RNAs as splicing factor hijackers. ( i) MALAT1 can disrupt the formation of a splicing modulator complex, by directly hijacking the SFPQ factor, thus inhibiting its interaction with the tumor growth factor PTBP2. SFPQ-released PTBP2 promotes the proliferation of cancer cells. (ii) Celf3 and SF1 normally co-localize in the CS nuclear bodies. GOMAFU lncRNA is recognized by both proteins, although it is not accumulated in CS bodies. It was proposed that GOMAFU promotes the re-localization of Celf and SF1 out from CS bodies to the nucleoplasm, modulating their activity in splicing. ( iii) SPA and sno-lncRNAs recruit RNA binding proteins such as FOX, TDP43 and hnRNP M, and may titrate their availability for splicing regulation throughout the nucleus. In PWS patients SPA and sno-lncRNAs loci are deleted or not expressed, thus the related proteins are more uniformly distributed, impacting the AS pattern of their target genes. (iv) ASCO lncRNA is directly recognized by NSRa and b, competing with their binding to NSR-targeted pre-mRNAs. ASCO-mediated modulation of NSRs results in alternative processing of mature mRNAs, exhibiting events of intron retention, exon skipping and alternative 5′ or 3′ ends. (v) ENOD40 lncRNA is directly recognized by the nuclear speckle protein RBP1. ENOD40 participates in the nucleocytoplasmic trafficking of RBP1, inducing its accumulation into cytoplasmic granules, likely modulating RBP1-dependent splicing.