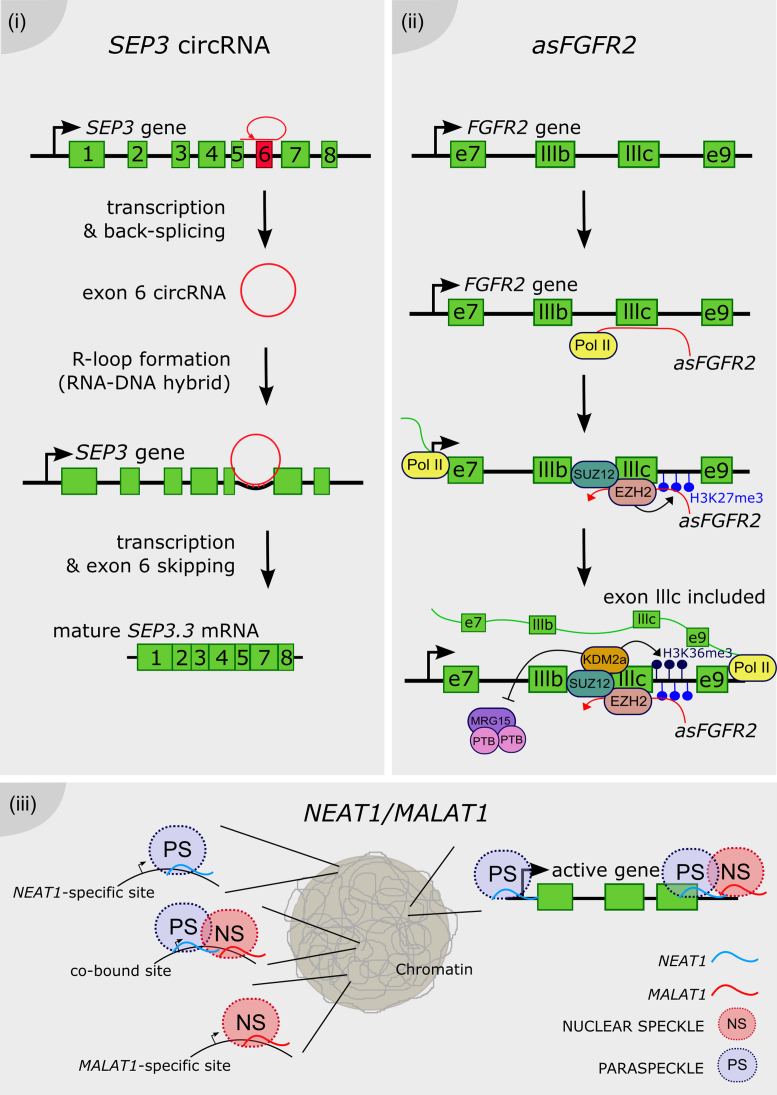
Figure 4.
Long noncoding RNAs as chromatin remodelers. (i) The exon 6 of the SEP3 gene is transcribed and back-spliced into a circular RNA. The SEP3 circRNA directly interacts with its parent gene DNA, conforming a DNA–RNA duplex known as an R-loop and promoting the exon 6 skipping, thus the accumulation of the SEP3.3 mRNA isoform. (ii) The antisense transcript of the FGFR2 gene, called asFGFR2 (in red), recruits the PRC2 proteins EZH2 and SUZ12 to its parent locus, triggering the deposition of H3K27me3 and the recruitment of the H3K36 demethylase KDM2a. This complex enhances the deposition of H3K36me3 and impairs the binding of the chromatin-splicing adaptor complex MRG15–PTB to the exon IIIb, which is finally included in the mature mRNA (in green). (iii) NEAT1 and MALAT1 bind to common and distinct actively transcribed loci across the genome. Their binding on the gene body is different between them, e.g. NEAT1 binding peaks at the transcription start site as well as the end of the locus, whereas MALAT1 preferentially binds only at the end of the gene. It was proposed that MALAT1 and NEAT1 promote the formation of splicing-related nuclear speckles and paraspeckles, respectively, around its site of transcription of targeted loci.