Abstract
As the most common type of DNA damage, DNA single-strand breaks (SSBs) are primarily repaired by the SSB repair mechanism. If not repaired properly or promptly, unrepaired SSBs lead to genome stability and have been implicated in cancer and neurodegenerative diseases. However, it remains unknown how unrepaired SSBs are recognized by DNA damage response (DDR) pathway, largely because of the lack of a feasible experimental system. Here, we demonstrate evidence showing that an ATR-dependent checkpoint signaling is activated by a defined plasmid-based site-specific SSB structure in Xenopus HSS (high-speed supernatant) system. Notably, the distinct SSB signaling requires APE2 and canonical checkpoint proteins, including ATR, ATRIP, TopBP1, Rad9 and Claspin. Importantly, the SSB-induced ATR DDR is essential for SSB repair. We and others show that APE2 interacts with PCNA via its PIP box and preferentially interacts with ssDNA via its C-terminus Zf–GRF domain, a conserved motif found in >100 proteins involved in DNA/RNA metabolism. Here, we identify a novel mode of APE2–PCNA interaction via APE2 Zf–GRF and PCNA C-terminus. Mechanistically, the APE2 Zf–GRF–PCNA interaction facilitates 3′-5′ SSB end resection, checkpoint protein complex assembly, and SSB-induced DDR pathway. Together, we propose that APE2 promotes ATR–Chk1 DDR pathway from a single-strand break.
INTRODUCTION
As the most common type of DNA damage in the genome, DNA single-strand breaks (SSBs) are generated from unbalanced reactive oxygen species, intermediate products of DNA repair pathways such as base excision repair (BER), and aborted activity of cellular enzymes such as Topoisomerase 1 (Top1) (1–3). SSBs are primarily resolved by a global SSB repair pathway, including recognition by sensor proteins such as PARP1, SSB processing by enzymes, gap filling through insertion or nucleotide extension, and ligation by DNA ligation enzymes such as DNA ligase I or IIIα (1,2,4). In addition, SSBs can also be resolved by homologue recombination or alternative homologue-mediated SSB repair (5). Defects in SSB repair have been associated with cancer and neurodegenerative disorders as well as genetic diseases such as ataxia-oculomotor apraxia 1 (AOA1) and spinocerebellar ataxia with axonal neuropathy 1 (SCAN1), underlining the significance of SSB repair (1,2). However, it remains unknown how unrepaired SSBs are sensed and signaled via DNA damage response (DDR) pathways.
Cellular response to DNA damage and replication stress is mainly coordinated by ATR–Chk1 and/or ATM-Chk2 DDR pathways (6,7). ATR–Chk1 DDR pathway can be activated in response to stalled DNA replication forks, UV-damage, DSBs or oxidative DNA damage (7–9). Full ATR activation requires several mediator proteins, such as ATRIP, TopBP1, MDC1 and the 9–1–1 (Rad9-Rad1-Hus1) complex (10–12). It is generally accepted that an RPA-coated long stretch of ssDNA serves as a platform to recruit checkpoint proteins to sites of DNA damage (13,14). Activated ATR kinase phosphorylates a variety of substrates including Chk1 to regulate cell cycle progress, activate transcription, and promote DNA repair (15). Chk1 phosphorylation is often utilized as an indicator of ATR activation (16). In response to DSBs, ATM is activated by auto-phosphorylation and monomer formation, leading to phosphorylation of its downstream substrates including Chk2 (17). DSB end resection in the 5′-3′ direction is proposed to couple DSB repair and ATM-Chk2 DDR pathway (18). A two-step model for DNA end resection at DSB sites has been suggested to occur through MRN (Mre11-Rad50-Nbs1) complex or CtIP/Sae2, and Exo1 or DNA2/Sgs1 (19). Furthermore, ATM can be activated through a disulfide bond formation and conformation change in oxidative stress in a DNA-independent manner (20,21). Although ATM can be activated by presumptive unrepaired SSBs in XRCC1-dificient cells, it remains unknown how exactly unrepaired SSBs activate ATM DDR pathway (21).
Several critical barriers hinder our understanding of SSB signaling at the molecular level. The first critical barrier is the lack of a defined experimental system to dissect all facets of SSB signaling. Cellular signaling to DSBs has been studied via generating a single site-specific DSB in genome by HO or I-SceI endonuclease in yeast and mammalian cells (22,23). Our understanding of SSB signaling is mostly from experimental systems using indirect SSBs after treatment of exogenous reagents, such as hydrogen peroxide and methyl methanesulfonate (MMS) (21,24). Additionally, spatial and temporal cellular response to multiple SSBs induced by UVDE (UV damage endonuclease) was partially characterized in human cells (25). However, it remains unknown whether a defined SSB structure triggers a DDR pathway. The second critical barrier is the inability to distinguish SSBs from DSBs. Many DNA damaging reagents generate both SSBs and DSBs simultaneously or sequentially. Thus, it is extremely difficult to directly explore SSB signaling in response to only SSBs, as opposed to a combination of SSBs and DSBs.
Whereas APE1 is the major AP endonuclease (26), APE2 (APEX2, APN2) has strong 3′-5′ exonuclease and 3′-phosphodiesterase activities but weak AP endonuclease activity (27). APE2 is necessary for normal B cell development and recovery from chemotherapy drug-induced DNA damage (28). The interdomain connector loop (IDCL) of PCNA associates with the PIP (PCNA interaction protein) box of its interacting proteins (29). The PIP box of APE2 is important for PCNA association (24,30,31). Additionally, APE2 is a key player in PCNA-dependent repair of hydrogen peroxide-induced oxidative DNA damage (30,31). We have demonstrated that ATR–Chk1 DDR pathway is activated by hydrogen peroxide-induced oxidative stress in Xenopus, and that APE2 is important for the oxidative stress-induced ATR–Chk1 checkpoint signaling (24). Notably, our new evidence suggests that a zinc-finger motif (designated as Zf–GRF) in APE2’s C-terminus associates with ssDNA, but not dsDNA, and that APE2 Zf–GRF facilitates 3′-5′ end resection of oxidative DNA damage to promote ATR–Chk1 DDR pathway (32).
As a cell-free experimental system from eggs of the African clawed frog, Xenopus egg extracts have been widely used in studies of chromosome metabolism, and findings from Xenopus system can be validated in mammalian cell lines (33–35). Three different types of Xenopus egg extracts have been commonly used: low-speed supernatant (LSS), high-speed supernatant (HSS), and nucleoplasmic extracts (NPE) (36–38). Here, we have developed a plasmid-based SSB structure at a defined location and found that an ATR-dependent but replication-independent DDR pathway is activated by the defined SSB structure in the Xenopus HSS system. The SSB signaling requires APE2 and canonical checkpoint proteins including ATR, ATRIP, TopBP1, Rad9, Claspin. Surprisingly, we found that APE2’s Zf–GRF associates with PCNA through its C-terminus. We demonstrate evidence that the distinct APE2–PCNA interaction plays an essential role for the 3′-5′ SSB end resection and SSB signaling in a eukaryotic system. In addition, our data suggest that the SSB-induced ATR activation is important for SSB repair and that hydrogen peroxide triggers ATR-dependent DDR pathway in human cultured cells.
MATERIALS AND METHODS
Experimental procedures for egg extracts and chromatin preparation, SSB signaling technology, and plasmid DNA-bound fraction isolation in Xenopus laevis
The care and use of Xenopus laevis was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Charlotte. Sperm chromatin was prepared and utilized according to methods as described previously (11,24,35,39). The preparation of Xenopus LSS, HSS and NPE were described previously (35,36,39). DNA synthesis analysis from the HSS/NPE system was performed as previously described (11,35). In general, immunodepletion of target protein in HSS was performed with a similar procedure for immunodepletion in LSS as previously described (24,32). For example, to deplete APE2 from HSS, 40 ul of HSS was incubated with ∼20 ul of ProteinA Sepharose beads (GE Healthcare), which was pre-coupled with 20 ul of anti-APE2 antiserum (24), for 30–40 min at 4°C with constant mixing. Typically, 3-round depletion is needed to get ∼20 ul of APE2-depleted HSS from 40 ul of HSS. Immunodepletion of other target proteins in HSS shown in Figure 3 was performed using a similar approach as APE2 depletion in HSS. Antibodies used for immunodepletion were kind gifts from Drs Howard Lindsay (ATRIP and Rad9), Matthew Michael (TopBP1) and Karlene Cimprich (Claspin), respectively (11,24,40,41). Antibodies against XRCC1 were raised in rabbits against recombinant GST-XRCC1 (Cocalico Biologicals, see below section for the preparation).
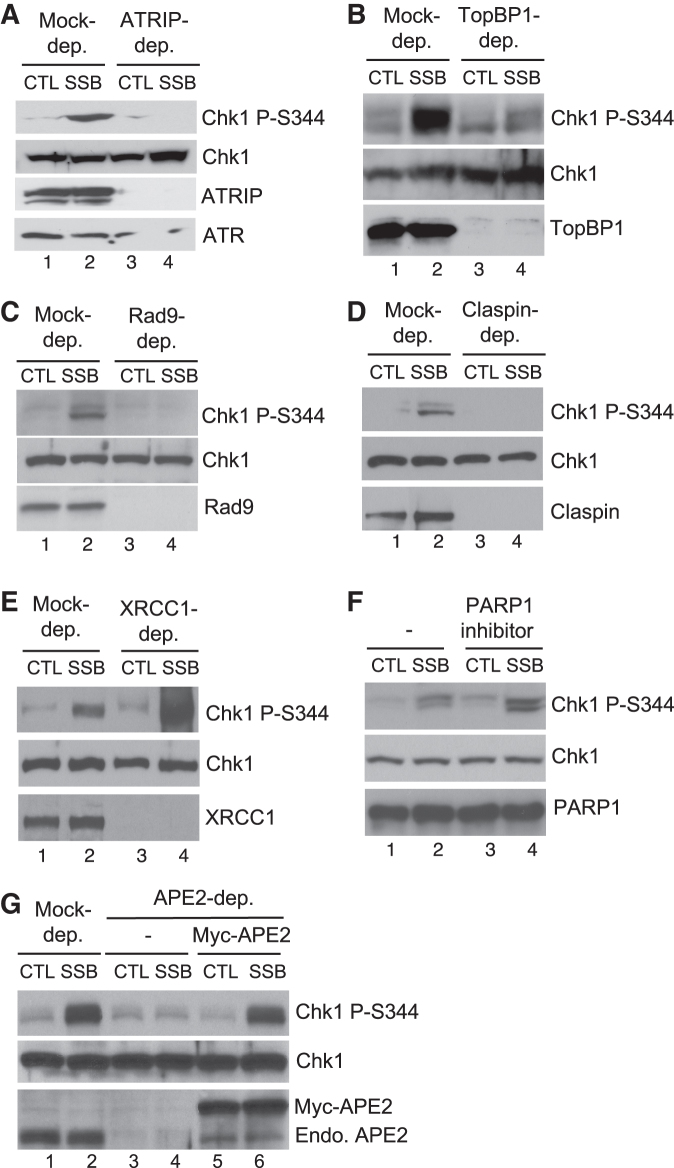
Figure 3.
APE2 is required for checkpoint signaling from the defined SSB structure in the HSS system. (A–E) CTL or SSB plasmid was added to mock-, ATRIP-, TopBP1-, Rad9-, Claspin-, or XRCC1-depleted HSS, respectively, at a concentration of 75 ng/μL for 30 min. Extracts were examined via immunoblotting analysis, as indicated. (F) PARP1 specific inhibitor (4-amino-1,8-naphthalimide, 0.1 mM) was added to HSS supplemented with CTL or SSB plasmid. Extracts were examined via immunoblotting analysis. (G) Recombinant Myc-APE2 was added to APE2-depleted HSS supplemented with CTL or SSB plasmid. Extracts were examined via immunoblotting analysis. ‘Endo. APE2’ represents endogenous APE2.
For the SSB signaling experiments, typically 8 ul of HSS was supplemented with 2 ul of either control or SSB plasmid (see below section for details) to a final concentration. After incubation of different time as indicated at room temperature, the 10 ul of reaction mixture (i.e., HSS-plasmid mixture) was added with 40 ul of sample buffer, which was examined via immunoblotting analysis (e.g., 10 ul of samples were loaded per lane).
For DNA-bound protein isolation from HSS system (Figure 6A), after room temperature incubation, 50 ul of reaction mixture was diluted with 200 ul of egg lysis buffer (ELB, 250 mM sucrose, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES, pH 7.7) and spin through a 1 ml of sucrose cushion (0.9 M sucrose, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES, pH 7.7) at 10 000 rpm for 15 min at 4°C with a swinging bucket. After centrifugation, the supernatants were removed, and the DNA-bound protein factions were resuspended with sample buffer and examined via immunoblotting analysis.
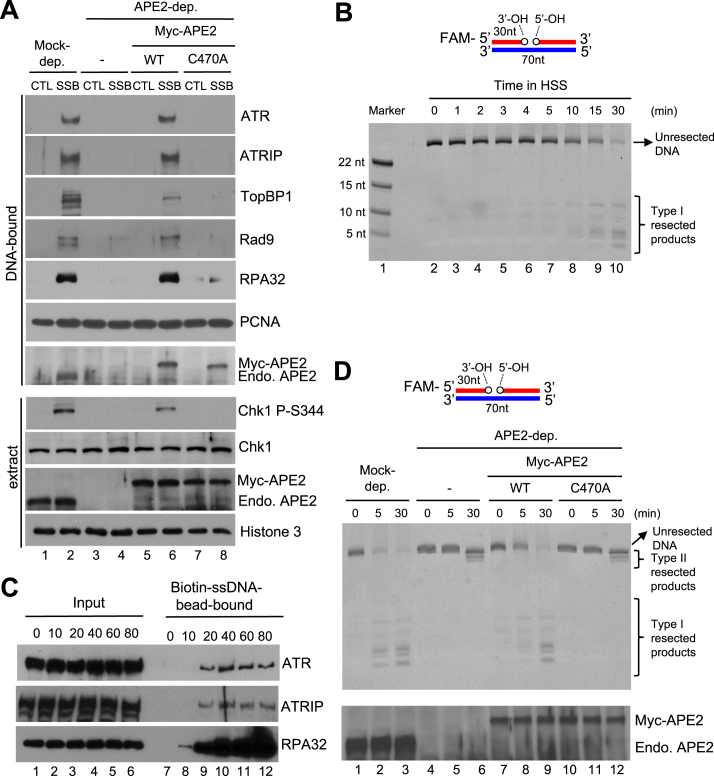
Figure 6.
APE2 Zf–GRF–PCNA interaction promotes SSB end resection, the assembly of a checkpoint protein complex onto SSB plasmid, and Chk1 phosphorylation in the HSS system. (A) CTL or SSB plasmid was added to mock- or APE2-depleted HSS, which was supplemented with WT or C470A Myc-APE2. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated. ‘Endo. APE2’ represents endogenous APE2. (B) FAM-labeled dsDNA with a site specific SSB (designed as FAM-SSB) was added to HSS for different time as indicated. Then samples were examined via TBE-Urea gel and visualized via Typhoon imager. ‘Marker’ represents four FAM-labeled different-length ssDNA. (C) The length dependence of ssDNA for the recruitment of ATR-ATRIP complex and RPA to ssDNA in the HSS. Streptavidin Dynabeads coupled with different length of Biotin-coupled ssDNA (i.e., 0, 10, 20, 40, 60 or 80nt) were added to HSS. After incubation, the Biotin-ssDNA bead-bound fractions were isolated from HSS. The Input and bead-bound fractions were examined via immunoblotting analysis. (D) The FAM-SSB substrate was added to mock- or APE2-depleted HSS, which was supplemented with WT or C470A Myc-APE2. DNA structures were examined via TBE-Urea gel and visualized via Typhoon imager (top). Samples were also analyzed via immunoblotting analysis as indicated. ‘Endo. APE2’ represents endogenous APE2 (bottom).
Preparation of SSB and DSB plasmid as well as FAM-SSB structure
There are four recognition sites on pUC19 for Nt.BstNBI, designated as site1 (nt 427–431 on (+) strand), site2 (nt 1177–1181 on (+) strand), site3 (nt 706–710 on (–) strand), and site4 (nt 1694–1698 on (–) strand). The plasmid pS was generated by mutant pUC19 on three sites (i.e. site2–site4) sequentially with three pairs of primers using QuikChange II XL site-directed mutagenesis kit. The mutations were verified and confirmed by DNA sequencing. Qiagen plasmid midi kit was utilized to obtain large amounts of the pS plasmid.
To generate a defined SSB between C435 and T436, the pS was treated with Nt. BstNBI (10 U/μg) for 2 h at 55°C and CIP (calf intestine phosphatase, 10 U/μg) for 1 h at 37°C to remove the 5′-P of T436. The SSB plasmid was purified from agarose via QIAquick gel extraction kit and then optionally purified by phenol–chloroform extraction. To generate a DSB plasmid, the pS was treated with SbfI-HF at 37°C and subsequently with CIP at 37°C. The DSB plasmid was purified from agarose via QIAquick gel extraction kit and then optionally purified by phenol–chloroform extraction.
A FAM-SSB structure was generated by PCR using the pS as template following by nicking enzyme treatment. The 70bp dsDNA PCR product (i.e. bp 406–475) was purified from agarose via QIAquick gel extraction kit and subsequently treated with Nt. BstNBI (10 U/μg) for 2 h at 55°C and CIP (10 U/μg) for 1 h at 37°C. The FAM-SSB structure was purified from agarose via QIAquick gel extraction kit and then purified by phenol–chloroform extraction.
Recombinant DNA and proteins
Recombinant pGEX-4T1-WT APE2-ZF was generated by cloning the ZF domain (nt 1478–1666) of xlAPE2 (GenBank: BC077433, Xenopus Gene Collection IMAGE ID: 4030411), which corresponds to the aa 456–517, into EcoRI- and XhoI-digested pGEX-4T1. Recombinant pET28a-PCNA was generated by subcloning full-length coding region (nt 39–824) of xlPCNA (GenBank: BC057758, Xenopus Gene Collection IMAGE ID:5049027) into BamHI- and NotI-digested pET28a. Recombinant pGEX-4T1-XRCC1 was generated by cloning the coding region (nt 164–2119) of xlXRCC1 (GenBank: BC045032, Xenopus Gene Collection IMAGE ID:5543195) into EcoRI- and XhoI-digested pGEX-4T1. Recombinant pGEX-4T1-APE1 was generated by cloning the coding region (nt 119–1069) of xlAPE1 (GenBank: BC072056, Xenopus Gene Collection IMAGE ID: 4202632) into BamHI- and XhoI-digested pGEX-4T1.
Point mutants of recombinant DNA were generated with QuikChange IIXL Site-Directed Mutagenesis kit (Agilent). Recombinant plasmids were made via QIAprep spin miniprep kit following vendor's standard protocol. Myc-tagged recombinant proteins were generated with various pCS2+MT-derived recombinant plasmids and TNT SP6 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's protocol. GST-or His-tagged recombinant proteins were expressed and purified in Escherichia coli DE3/BL21 following vendor's standard protocol. Purified recombinant proteins were confirmed on coomassie-stained SDS-PAGE gels with a range of BSA standards and a pre-stained protein ladder.
Immunoblotting analysis and antibodies
Anti-XRCC1 antibodies were raised in rabbits against GST-XRCC1 (Cocalico Biologicals). Anti-Xenopus APE2 antibodies were described previously (24). Antibodies against ATR and Claspin were provided by Dr. Karlene Cimprich (33,41). Antibodies against ATRIP, Rad9, and Rad17 were provided by Dr Howard Lindsay (40). Antibodies against TopBP1 and RPA32 were provided by Dr Matthew Michael (11). Antibodies against PARP1 were provided from Dr Yoshiaki Azuma (43). Antibodies against human APE2 were provided by Drs Yusaku Nakabeppu and Daisuke Tsuchimoto (44). Antibodies against RPA32 phosphorylation at Ser33 and Rad17 phosphorylation at Ser645 were purchased from Bethyl Laboratories. Antibodies against Chk1 phosphorylation at Ser345 were purchased from Cell Signaling Technology. Antibodies against Histone 3 were purchased from Abcam. Antibodies against Chk1, GST, His, Myc, PCNA and Tubulin were purchased from Santa Cruz Biotechnology. Antibodies against human Chk1 and human RPA32 were purchased from Cell Signaling Technology and Thermo Scientific, respectively.
GST pulldown assays
For the GST-pull-down experiments from HSS, 5 μg of GST or GST-tagged recombinant proteins were added to 200 μl Xenopus HSS. After an hour of incubation, an aliquot of egg extract mixture was collected as Input and the remaining extract mixture was diluted with 200 μl Interaction Buffer (100 mM NaCl, 5 mM MgCl2, 10% (vol/vol) glycerol, 0.1% Nonidet P-40, 20 mM Tris–HCl at pH 8.0). Then, 30 μl of glutathione beads that were resuspended in 200 μl interaction buffer were added to the diluted egg extracts. After 1 h incubation at room temperature, beads were washed with Interaction Buffer two times. Then, the bead-bound fractions and Input were analyzed via immunoblotting.
For the GST-pull-down experiment from a buffer, 5 μg of GST or GST-tagged recombinant proteins, and 5 μg of WT or mutant His-tagged PCNA were added to 200 μl Interaction Buffer. After an hour of incubation, an aliquot of the mixture was collected as Input and the remaining mixture was supplemented with 100 μl interaction buffer that contains 30 μl glutathione beads. After 1 h incubation at room temperature, beads were washed with Interaction Buffer two times. Then, the bead-bound fractions and the Input were analyzed via immunoblotting.
Analysis of DNA repair products from the HSS system
Nuclease-free water was added to each reaction of HSS that was incubated with SSB or DSB plasmid to a total of 300 μl. Equal volume of phenol-chloroform was added to the mixture and resuspended up and down five times and spin for 5 min at 13 000 rpm at room temperature. The top aqueous layer was transferred to a new tube and the phenol–chloroform extraction was repeated. Then, sodium acetate (pH 5.0, a final concentration of 0.3 M) and glycogen (a final concentration of 1 μg/μl), as well as ethanol (100%, 2.5-fold volume), were added to the aqueous solution, which was incubated for 30 min at –70°C. The mixture was centrifuged for 15 min at 13 000 rpm at room temperature. The pellet was washed by cold 70% ethanol and air-dried for 30 min before resuspension with nuclease-free water. Then the purified DNA repair products were analyzed on agarose gel and stained with ethidium bromide.
SSB end resection assays in the HSS system
The FAM-SSB structure was added to mock- or APE2-depleted HSS, which was supplemented with WT/mutant Myc-tagged APE2, respectively. After different times of incubation at room temperature, reactions were quenched with equal volume of TBE-Urea Sample Buffer (Invitrogen), denatured at 95°C for 5 min, and cooled at 4°C for 2 min. Samples were examined on 20% TBE-Urea gel and imaged on Typhoon 8600 and viewed using ImageQuant software.
In vitro exonuclease assays
Previous biochemistry analysis has indicated that APE1 can resect nicked dsDNA into 1–3nt gap in the 3′-5′ direction in vitro (45). To generate a FAM-labeled gapped structure, we found that our recombinant GST-APE1 resected FAM-labeled SSB substrate in the 3′-5′ direction in a dose-dependent manner (Supplementary Figure S5A). Thus, we utilized this APE1-pretreated FAM-labeled gapped substrate for our APE2 exonuclease analysis in vitro. For the in vitro exonuclease assays, the FAM-SSB substrate was pretreated with recombinant APE1 in exonuclease buffer (20 mM KCl, 10 mM MCl2, 2 mM DTT, 50 mM HEPES, pH 7.5) at 95°C for 20 min, followed by phenol-chloroform extraction and purification. This APE1-treatment method is derived and modified from a method described previously (45). The purified gapped dsDNA structure (50 nM) was incubated in 1× reaction buffer (50 mM NaCl, 1 mM TCEP, 1 mM MnCl2, 10 mM Tris–HCl, pH 8.0) with different combinations of purified recombinant proteins. After a 30-min incubation at 37°C, reactions were quenched with equal volume of TBE–Urea Sample Buffer, denatured at 95°C for 5 min, and cooled at 4°C for 2 min. Samples were loaded and resolved on a 20% TBE–Urea gel. Gels were imaged using a Typhoon imager (GE Healthcare) and viewed using ImageJ.
DNA binding assays
For the ssDNA binding assays in a buffer using GST or GST-tagged proteins, 40 μl of biotin–ssDNA (80nt, 100 μM) was added to 40 μl of Streptavidin Dynabeads in 2× B&W Buffer (2 M NaCl, 1 mM EDTA, 10 mM Tris–HCl, pH 7.5), and incubated for 15 min at room temperature. After separating beads from buffer, the beads were washed by 2× B&W Buffer for three times and resuspended in 100 μl of Buffer B (80 mM NaCl, 20 mM β-glycerophosphate, 2.5 mM EGTA, 0.01% NP-40, 10 mM MgCl2, 100 ug/ml BSA, 10 mM DTT and 10 mM HEPES–KOH, pH 7.5). Then 20 μg of GST or GST-tagged proteins in 100 μl of Buffer B was added to the 80nt-ssDNA-coupled beads in 100 μl of Buffer B. After a 30-min incubation at 4°C, an aliquot of mixture was collected as Input, and the beads were washed for three times with Buffer A (80 mM NaCl, 20 mM β-glycerophosphate, 2.5 mM EGTA, 0.01% NP-40 and 10 mM HEPES–KOH, pH 7.5). The Input and Bead-bound fractions were examined via immunoblotting analysis.
For the ssDNA binding assays in the HSS system, different lengths of Biotin-ssDNA (i.e. 0, 10, 20, 40, 60 and 80) were coupled to Streptavidin Dynabeads using a similar approach as above described, and resuspended in 200 μl of Buffer B. Then, the 200 μl of beads coupled with ssDNA was added to equal volume of diluted HSS (1:1 v/v dilution with Buffer B). After 1-h incubation, an aliquot of the mixture was collected as Input, and the beads were washed with Buffer A three times. The Input and Bead-bound fractions were examined via immunoblotting analysis.
For the gapped DNA binding assays with purified proteins, biotin-gapped DNA structure was prepared in a similar approach to that for the FAM-gapped DNA structure with the exception that biotin was covalently linked to the 5′ side of one primer. The coupling of biotin-gapped DNA structure to streptavidin dynabeads, protein incubation and bead washing process were performed following the protocol for ssDNA binding analysis in a buffer as described above.
Cell culture, treatment and analysis
Human U2OS cells were purchased from the American Type Culture Collection (ATCC) and cultured in Dulbecco's Modified Eagle Medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS, Invitrogen), penicillin (100 U/ml) and streptomycin (100 μg/ml, Life Technologies) in incubator at 37°C and 5% CO2. For G1 synchronization, cells were first blocked in S phase with 2 mM thymidine for 24 h and released for 4 hours, then blocked in M phase with 100 ng/ml nocodazole for 12 h and released for 6 h, and finally blocked in G1 phase with DMEM without FBS for 96 h. Cells were treated with VE-822 (5 μM) in culture medium for 1 h followed by hydrogen peroxide treatment (1.25 mM) for 4 h. Then cells were collected and split for immunoblotting analysis and FACS analysis. For immunoblotting analysis, equal total proteins of cell lysates (10 μg per lane) were loaded and Tubulin was used for loading control. For FACS analysis, cells were further fixed and stained with DAPI followed by cell cycle profiling using FACS machine (BD LSRFortesssa) following manufacturer's standard protocol.
Quantification
ImageQuant software was utilized to quantify the incorporation of [32P-α]-dATP to newly synthesized DNA from the HSS/NPE system. ImageQuant was used to visualize gels from SSB end resection assays. ImageJ was utilized to view gels from exonuclease assays and to quantify SSB repair products on agarose.
RESULTS
A defined SSB structure can be repaired in the HSS system
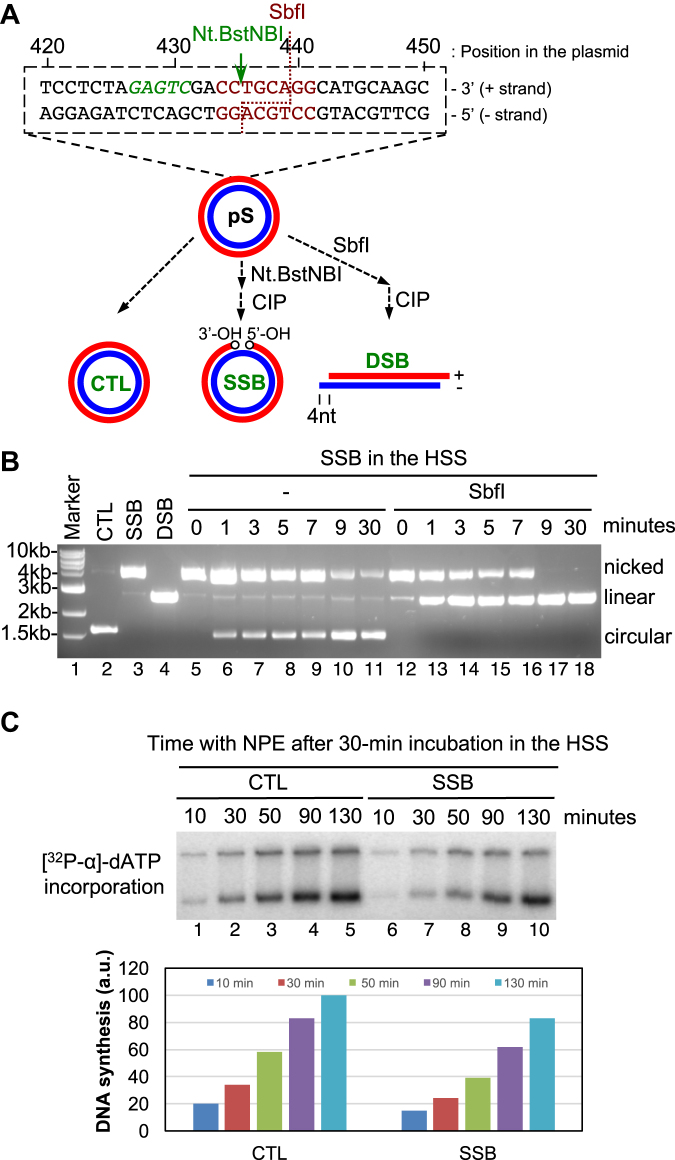
We generated a pUC19-derived plasmid pS that contains only one recognition sequence for a nicking endonuclease Nt.BstNBI (Figure 1A). pS was treated by Nt.BstNBI and calf intestinal alkaline phosphatase (CIP) sequentially to generate a SSB structure containing only one nick between dC435 and dT436 in the ‘+’ strand with 3′-OH and 5′-OH at both ends. pS was treated with restriction enzyme SbfI and CIP sequentially to generate a DSB structure (Figure 1A). The SSB, DSB and CTL (control) plasmid structures are distinguished on agarose gel based on their structure-dependent mobility shift (Figure 1A and Supplementary Figure S1A). Because the nick is exactly in the recognition sequence of SbfI, the SSB structure was resistant to further SbfI treatment, as expected (Supplementary Figure S1A).
Figure 1.
A defined site-specific SSB structure can be repaired in the Xenopus HSS system. (A) Schematic diagram of generating SSB and DSB plasmid structures. (B) The defined SSB structure is gradually repaired in HSS system. SSB repair intermediate products from HSS system were isolated at different time points and treated by SbfI, followed by examination via agarose gel electrophoresis. (C) (Top) CTL or SSB plasmid was added to HSS, which was supplemented with [32P-α]-dATP, for a 30-min incubation. Then NPE was added for continuous incubation for different time as indicated and samples were examined on agarose gel. (Bottom) Quantification of DNA synthesis of CTL or SSB plasmid in the HSS/NPE system shown in the top.
To examine SSB repair process in the HSS system, we found that the nicked version was converted into a circular version after different time incubation in HSS, indicating that the SSB structure is gradually repaired in HSS (lanes 6–11, Figure 1B). Moreover, the circular repair products of SSB structure from HSS were catalyzed by SbfI into a linear version (lanes 12–18, Figure 1B). Strikingly, a portion of the nicked version of SSB structure developed SbfI sensitivity at beginning time points (1, 3, 5 and 7 min), whereas all SSB structures were sensitive to SbfI at late time points (9 and 30 min) (Figure 1B). These observations suggest that, after HSS incubation, the defined SSB structure is either repaired into a circular version or converted into an intermediate product with a normal SbfI recognition sequence but with a nick and/or gap at another location. As expected, the CTL plasmid isolated from the HSS was catalyzed by SbfI into linear versions at all time points (Supplementary Figure S1B). Circular plasmid DNA can be assembled into pre-Replication Complex in the HSS, but cannot continue its DNA synthesis without addition of the NPE supplying S-phase CDKs (37). To confirm the defined SSB structure is indeed repaired in the HSS, we examined them using the Xenopus HSS/NPE system (37). Similar to CTL plasmid, the defined SSB structure can be replicated efficiently in the HSS/NPE system (only ∼17% reduction), suggesting that most SSB structures have been repaired after a 30-min HSS incubation (Figure 1C).
A distinct ATR–Chk1 DNA damage response pathway is induced by a defined SSB structure in the HSS system
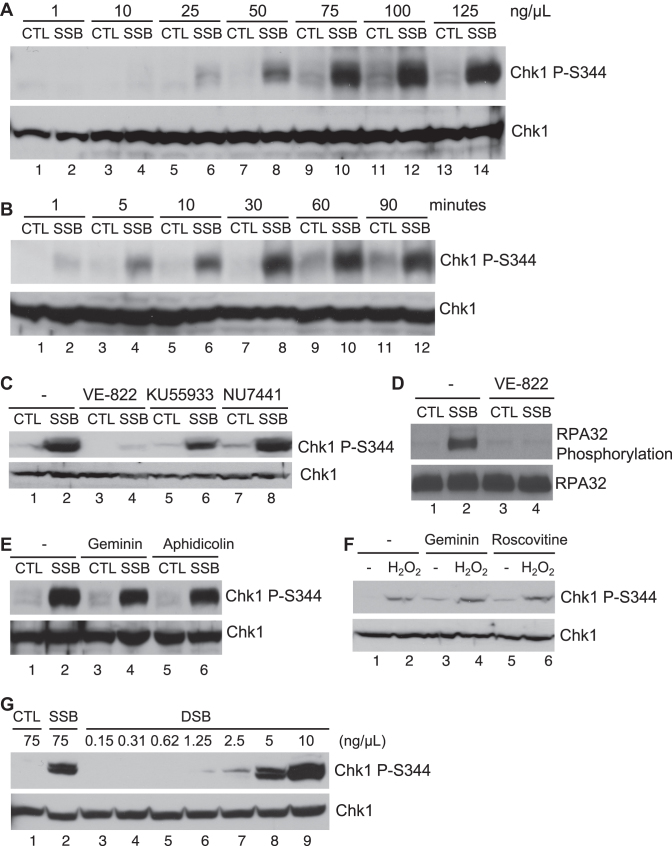
To determine whether a defined SSB structure triggers a DDR pathway, we added different concentrations of SSB or CTL plasmid structure to HSS, followed by immunoblotting analysis after a 30-min incubation. We revealed that 75 ng/μl (43 nM) of the defined SSB plasmid, but not CTL plasmid, triggered a robust Chk1 phosphorylation at S344 in the HSS (equivalent to Chk1 phosphorylation at S345 in humans), suggesting that the SSB structure induces a unique checkpoint signaling in the HSS in a dose-dependent manner (Figure 2A). Our time-dependence analysis found that the SSB-induced Chk1 phosphorylation appeared at 5 minutes, and peaked at 30 min after incubation in the HSS (Figure 2B). Whereas ATM specific inhibitor KU55933 and DNA-PK specific inhibitor NU7441 almost had no effect on SSB-induced Chk1 phosphorylation, the addition of ATR specific inhibitor VE-822 compromised the SSB-induced Chk1 phosphorylation, suggesting that the SSB signaling is ATR-dependent (Figure 2C). In addition to Chk1, RPA32 and Rad17 are substrates of ATR kinase (46–49). Consistent with Chk1 phosphorylation, RPA32 phosphorylation and Rad17 phosphorylation were induced by the defined SSB in the HSS in a VE-822 sensitive manner (Figure 2D and Supplementary Figure S2A). In addition, we verified that potential DSB contamination in our SSB plasmid is less than ∼1% by quantification. The defined DSB structure required at least 5 ng/μl (3 nM) to trigger a robust Chk1 phosphorylation in the HSS, suggesting that the defined SSB-induced Chk1 phosphorylation is not due to confounding DSB in our SSB preparation (Figure 2G).
Figure 2.
An ATR–Chk1 DNA damage response pathway is induced by the defined SSB structure in the HSS system. (A) CTL or SSB plasmid was added to HSS at different concentrations as indicated, and incubated for 30 min. Extracts were examined via immunoblotting analysis for Chk1 phosphorylation (i.e., Chk1 P-Ser344) and total Chk1. (B) CTL or SSB plasmid was added to HSS at a final concentration of 75 ng/μl. After different time of incubation at room temperature, the extracts were examined via immunoblotting analysis. (C–E) ATR inhibitor VE-822, ATM inhibitor KU55933, DNA-PK inhibitor NU7441, recombinant geminin, or DNA polymerase inhibitor Aphidicolin was added to HSS supplemented with CTL or SSB plasmid at a final concentration of 75 ng/μl for 30 min. Extracts were examined via immunoblotting analysis as indicated. (F) Geminin or roscovitine was added to HSS supplemented with sperm chromatin and hydrogen peroxide. After a 45-min incubation, extracts were examined via immunoblotting analysis as indicated. (G) CTL, SSB or DSB plasmid was added to HSS at different concentrations as indicated. Samples were examined via immunoblotting analysis.
Although DNA replication is required for ATR–Chk1 checkpoint activation in response to stalled replication forks and oxidative stress (32,33,50), the defined SSB-induced Chk1 phosphorylation was not impaired by the addition of MCM helicase inhibitor recombinant geminin protein, suggesting that DNA replication is dispensable for the SSB-induced Chk1 phosphorylation in the HSS system (Figure 2E). This observation is consistent with the deficiency of plasmid DNA replication elongation in Xenopus HSS without addition of NPE (37). Aphidicolin, a small molecule inhibitor of DNA polymerase alpha, delta, and epsilon, can induce stalled DNA replication forks that trigger ATR activation in the Xenopus LSS system (11,46,47). However, we found that the SSB-induced Chk1 phosphorylation was not affected by the addition of aphidicolin (Figure 2E). Furthermore, the SSB-induced Chk1 phosphorylation was not affected when Pol alpha (p70 subunit) was depleted in the HSS system (Supplementary Figure S2C). This feature of the SSB-induced replication-independent Chk1 phosphorylation in the HSS system is similar to the DSB-mimicking structure AT70-induced Chk1 phosphorylation in a replication-independent manner in the LSS system (34,51). Moreover, Chk1 phosphorylation was triggered by hydrogen peroxide when HSS was supplemented with sperm chromatin; however, the addition of geminin and CDK inhibitor rescovitine had no effect on the hydrogen peroxide-induced Chk1 phosphorylation in the HSS (Figure 2F).
To further determine whether the defined SSB-induced Chk1 phosphorylation is ATR-dependent, we removed endogenous ATRIP via immunodepletion using anti-ATRIP antibodies which also co-depleted endogenous ATR, as ATR and ATRIP form a tight complex in Xenopus egg extracts (Figure 3A) (24). Notably, the SSB-induced Chk1 phosphorylation was compromised when ATRIP and most of ATR were absent in the HSS system (Figure 3A). This observation strongly suggests that the defined SSB structure triggers ATR activation. Together, the above observations suggest that a defined site-specific SSB structure triggers ATR–Chk1 DDR pathway activation in a dose- and time-dependent, but replication-independent, fashion in the Xenopus HSS system.
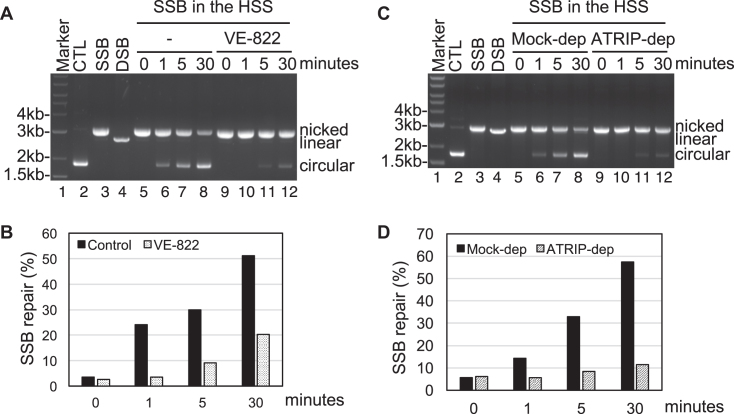
To identify the significance of SSB-induced ATR activation, we tested whether SSB repair is affected by ATR in the HSS system. We found that the addition of ATR specific inhibitor VE-822 compromised the SSB repair in the HSS system (Figure 4A and B). This observation strongly suggests that the SSB-induced ATR activation is important for SSB repair. To rule out the potential off-targets of VE-822, we sought to test whether SSB repair is impaired in ATRIP-depleted HSS. Consistent with the SSB repair impairment induced by ATR inhibitor VE-822 (Figure 4A and B), SSB repair capacity was compromised in ATRIP-depleted HSS (Figure 4C and D). Because anti-ATRIP antibodies co-depletes most ATR in HSS (see Figure 3A), the impairment of SSB repair in ATRIP-depleted HSS suggests that ATR-ATRIP DDR is essential for SSB repair. To address the physiological relevance of the SSB-induced ATR DDR pathway, we sought to test whether hydrogen peroxide induces ATR–Chk1 DDR in mammalian cells. A recent study using human U2OS cells demonstrated that ATR kinase is activated in G1 phase to facilitate the repair of ionizing radiation-induced DNA damage (52). Notably, we found that hydrogen peroxide triggers Chk1 phosphorylation and RPA32 phosphorylation in asynchronized U2OS cells (lanes 1 and 2, Supplementary Figure S3A). Importantly, addition of ATR specific inhibitor VE-822 prevented the hydrogen peroxide-induced Chk1 phosphorylation and RPA32 phosphorylation in asynchronized U2OS cells (lanes 3 and 4, Supplementary Figure S3A). We also observed that hydrogen peroxide induced ATR–Chk1 DDR in G1 synchronized U2OS cells (Supplementary Figure S3B). It seems that the potential effects on cell cycle profiling by H2O2 and VE-822 are minimal, if there is any (Supplementary Figure S3C). These observations in human U2OS cells suggest that our observations in Xenopus egg extracts system are very likely conserved in humans, demonstrating the physiological relevance of our research using Xenopus HSS system.
Figure 4.
The SSB-induced ATR DDR pathway is essential for SSB repair. (A) SSB plasmid was added to HSS with or without VE-822 for different time as indicated. SSB repair products were isolated and examined on agarose gel (Ethidium bromide staining). (B) Quantification of SSB repair capacity (circular/(circular + nicked) × 100) with or without VE-822 treatment in the HSS system from (A). (C) SSB plasmid was added to mock- or ATRIP-depleted HSS for different time as indicated. SSB repair products were isolated and examined on agarose (Ethidium bromide staining). (D) Quantification of SSB repair capacity in mock- or ATRIP-depleted HSS system from (C) using a similar method described in (B).
APE2 is required for the defined SSB-induced DDR pathway activation
Previous studies have shown that TopBP1, Rad9 and Claspin are canonical checkpoint proteins required for the ATR–Chk1 DDR pathway (8,34,42). Notably, the defined SSB-induced Chk1 phosphorylation was compromised when TopBP1, Rad9 or Claspin was immunodepleted in the HSS, respectively, suggesting the requirement of these checkpoint proteins for SSB signaling (Figure 3B–D). XRCC1 and PARP1 have been demonstrated in an association with SSB repair (43,53,54). We found that the defined SSB-induced Chk1 phosphorylation was enhanced when XRCC1 was immunodepleted in HSS or when PARP1 specific inhibitor (4-Amino-1,8-naphthalimide) was added to HSS, respectively (Figure 3E and F). Our interpretation of these observations is that the SSB signaling is enhanced when SSB repair is impaired by XRCC1 depletion or the addition of PARP1 inhibitor. Notably, the defined SSB-induced Chk1 phosphorylation is compromised in APE2-depleted HSS (Figure 3G). Importantly, recombinant WT Myc-tagged APE2 protein rescued the deficiency of Chk1 phosphorylation in APE2-depleted HSS (Figure 3G). These observations suggest that APE2 is required for the defined SSB-induced Chk1 phosphorylation in the HSS system.
APE2 Zf–GRF associates with PCNA C-terminal motif as a distinct mode of APE2–PCNA interaction
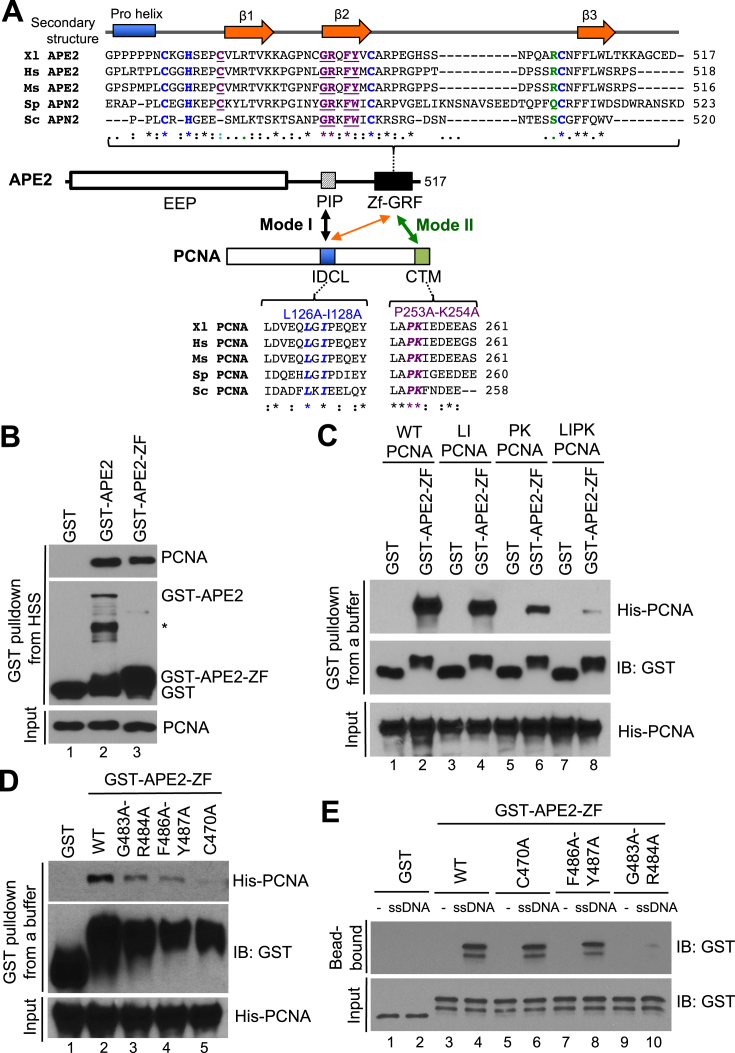
APE2 interacts with PCNA’s IDCL motif via its PIP box in yeast, Xenopus, and humans (Figure 5A) (24,30,31). Consistent with previous studies, our GST-pulldown assays demonstrated that PCNA associated with GST-APE2, but not GST, from Xenopus HSS, suggesting that APE2 associates with PCNA in the HSS (Figure 5B). Surprisingly, PCNA was also pulled down in the HSS by GST-APE2-ZF, which does not contain the PIP box, suggesting that APE2 Zf–GRF associates with PCNA in a PIP box-independent manner (Figure 5B). To further confirm the interaction between APE2 Zf–GRF with PCNA, we did GST-pulldown assays with recombinant PCNA protein and found that both GST-APE2 and GST-APE2-ZF, but not GST, pulldown recombinant PCNA in an interaction buffer, suggesting a possible direct interaction between APE2 Zf–GRF and PCNA (Supplementary Figure S4A). Notably, interaction with APE2 Zf–GRF was compromised in PK PCNA (P253A-K254A PCNA) and almost completely prevented in LIPK PCNA (L126A-I128A-P253A-K254A PCNA), suggesting that the PCNA C-terminal motif (CTM) plays an important role in Zf–GRF association (Figure 5A and C). To identify critical residues within APE2 Zf–GRF for its interaction with PCNA, we generated three point mutants in GST-APE2-ZF (i.e., G483A-R484A, F486A-Y487A and C470A). GST-pulldown assays demonstrated that G483A-R484A, F486A-Y487A, and C470A GST-APE2-ZF failed to efficiently interact with recombinant PCNA in an interaction buffer, in comparison to WT GST-APE2-ZF (Figure 5D). Because APE2 Zf–GRF associates with ssDNA (32), we intended to distinguish its interaction with PCNA from its association with ssDNA. We coupled 80nt ssDNA tagged with Biotin in the 5′ side to streptavidin beads, and found that WT GST-APE2-ZF, but not GST, interacts with ssDNA, as expected (Figure 5E). Importantly, G483A-R484A APE2-Zf–GRF is deficient in ssDNA interaction, whereas F486A-Y487A and C470A APE2-Zf–GRF are proficient for ssDNA binding (Figure 5E). In addition, R502E APE2 Zf–GRF is proficient in PCNA interaction although R502E APE2 Zf–GRF is deficient for ssDNA interaction and its exonuclease activity (32) (Supplementary Figure S4B and S4C). These observations suggest that APE2 Zf–GRF interaction with PCNA CTM is distinguished from its interaction with ssDNA. As the interaction of APE2 PIP box with PCNA IDCL motif is the first mode of APE2–PCNA interaction, we designated the APE2 Zf–GRF interaction with PCNA CTM as the second distinct mode of APE2–PCNA interaction (Figure 5A).
Figure 5.
APE2 Zf–GRF interacts with PCNA as the second mode of APE2–PCNA interaction. (A) Schematic diagram of APE2 Zf–GRF region and the IDCL and CTM regions of PCNA. (B) GST pulldown assays with GST, GST-APE2 and GST-APE2-ZF from HSS. The input and pulldown samples were examined via immunoblotting analysis. (C) GST pulldown assays with GST or GST-APE2-ZF as well as WT/mutant PCNA (i.e., LI PCNA, PK PCNA, or LIPK PCNA) in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (D) GST pulldown assays with GST or WT/mutant GST-APE2-ZF (i.e., G483A-R484A, F486A-Y487A, or C470A) as well as WT His-tagged PCNA in an interaction buffer. The input and pulldown samples were examined via immunoblotting analysis. (E) Biotin-coupled ssDNA (80nt) was coupled to streptavidin dynabeads and utilized for protein–DNA interaction assays with GST or WT/mutant GST-APE2-ZF (i.e., G483A-R484A, F486A-Y487A or C470A) in an interaction buffer. The input and bead-bound fractions were analyzed via immunoblotting analysis.
APE2 Zf–GRF–PCNA CTM interaction is critical for 3′-5′ SSB end resection, assembly of a checkpoint protein complex to SSB sites, and SSB signaling
To characterize the biological significance of APE2 Zf–GRF interaction with PCNA CTM motif, we added back WT or C470A Myc-tagged APE2 to APE2-depleted HSS and found that WT, but not C470A, APE2 rescued the SSB-induced Chk1 phosphorylation in APE2-depleted HSS, suggesting that the APE2 Zf–GRF–PCNA CTM interaction is important for the SSB-induced ATR–Chk1 DDR pathway in the HSS system (Figure 6A). Moreover, G483A-R484A APE2, which is deficient in interaction with ssDNA and PCNA CTM motif (Figure 5D and E), failed to rescue the SSB-induced Chk1 phosphorylation in APE2-depleted HSS (Supplementary Figure S4D). Furthermore, WT APE2, but not G483A-R484A APE2, rescued the hydrogen peroxide-induced Chk1 phosphorylation in APE2-depleted LSS system (Supplementary Figure S4E). These observations suggest that APE2 Zf–GRF interaction with PCNA CTM is important for SSB signaling.
Next, we isolated the DNA-bound fractions from HSS and examined the abundance of checkpoint proteins via immunoblotting analysis. Although PCNA was recruited to both CTL and SSB plasmid, a checkpoint protein complex, including ATR, ATRIP, TopBP1 and Rad9, was assembled onto SSB plasmid, but not CTL plasmid, in the HSS system (DNA-bound fractions, Lane 1–2, Figure 6A). Notably, APE2 preferentially associated with SSB plasmid, but not CTL plasmid, and RPA32 was also hyperloaded to SSB plasmid, but not CTL plasmid, in the HSS system, suggesting that the SSB plasmid is resected by APE2 into ssDNA for RPA binding and the assembly of the checkpoint protein complex (lanes 1–2, Figure 6A). When APE2 was removed via immunodepletion, the recruitment of RPA32, ATR, ATRIP, TopBP1 and Rad9 to SSB plasmid was compromised, further supporting the critical role of APE2 in the SSB end resection (lanes 3–4, Figure 6A). Importantly, WT APE2, but not C470A APE2, rescued the recruitment of RPA32, ATR, ATRIP, TopBP1 and Rad9 to SSB in APE2-depleted HSS (lanes 5–8, Figure 6A). Of note, similar to WT APE2, C470A APE2 was also recruited to SSB site efficiently in HSS, suggesting that APE2 PIP box interaction with PCNA IDCL motif is sufficient for the recruitment of APE2 to SSB site (Figure 6A). Together, our evidence demonstrates that the APE2 Zf–GRF interaction with PCNA CTM is critical for the checkpoint protein complex assembly onto SSB site and the SSB-induced ATR–Chk1 DDR pathway activation, but is dispensable for APE2 recruitment to SSB sites, in the HSS system.
To further investigate the SSB end resection by APE2 per se, we generated a FAM-labeled dsDNA in which a SSB is localized at a defined location in the upper strand, designated as FAM-SSB (70 bp in total, Figure 6B). After FAM-SSB was incubated in HSS for different time points, samples were examined via urea-denaturing PAGE electrophoresis. The FAM-SSB was resected in the 3′ to 5′ direction into Type I resected products in HSS (Figure 6B). Because the resected products are arranged from ∼4nt to ∼12nt, we estimated that the SSB structure is resected ∼18nt to 26nt in the 3′ to 5′ direction in the HSS system. Importantly, the 3′-5′ end resection of FAM-SSB was significantly compromised when APE2 was removed in HSS (top panel, lanes 1–6, Figure 6D). Interestingly, the FAM-SSB was still resected only 1nt–3nt, designated as Type II resected products, in APE2-depleted HSS (top panel, lane 6, Figure 6D). Although WT APE2 and C470A APE2 are added to similar concentrations in APE2-depleted HSS (bottom panel, Figure 6D), WT APE2 but not C470A APE2 rescued the deficiency of SSB end resection of FAM-SSB in APE2-depleted HSS (top panel, Figure 6D), suggesting that the APE2 Zf–GRF–PCNA CTM interaction is critical for the 3′-5′ SSB end resection in the HSS system.
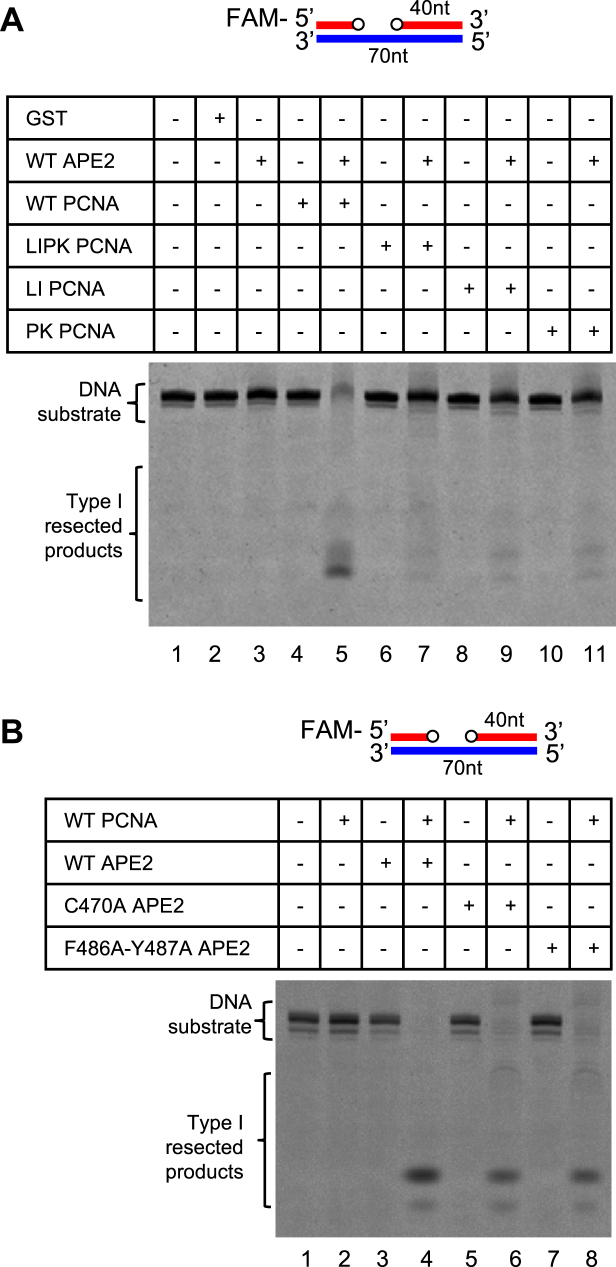
Using reconstitution system with purified proteins in vitro, we next examined DNA end resection of a gapped dsDNA structure, in which the FAM-SSB was pretreated with recombinant GST-APE1 to generate approximately 1nt–3nt gap (Supplementary Figure S5A). The gapped dsDNA was catalyzed into Type I resected products by recombinant GST-APE2 with the presence of WT, but not LIPK, LI, or PK, His-tagged PCNA, suggesting that IDCL and CTM regions within PCNA are all important for APE2’s exonuclease activity in vitro (Figure 7A). Surprisingly, the gapped dsDNA structure was still resected by C470A or F486A-Y487A GST-APE2 to some extent similar to WT GST-APE2 with the presence of WT His-tagged PCNA (Figure 7B). The different requirement of APE2 Zf–GRF interaction with PCNA CTM for SSB end resection in the HSS and in purified protein system in vitro may be because of a previously unidentified negative regulatory factor in the HSS system. We speculate that the APE2 Zf–GRF–PCNA CTM interaction is needed to overcome such inhibition of SSB end resection in the HSS system.
Figure 7.
Exonuclease activities of APE2 with purified proteins in vitro. (A) In vitro analysis of exonuclease activity of GST-APE2 with the presence or absence of WT or mutant His-tagged PCNA using the FAM-labeled gapped dsDNA structure. (B) In vitro analysis of exonuclease activity of WT and mutant GST-APE2 with the presence or absence of WT His-tagged PCNA using the FAM-labeled gapped dsDNA structure.
Together, our data suggest that the APE2 Zf–GRF–PCNA interaction promotes the ATR–Chk1 DDR pathway activation from a site-specific SSB structure in a cell-free eukaryotic system (Figure 8).
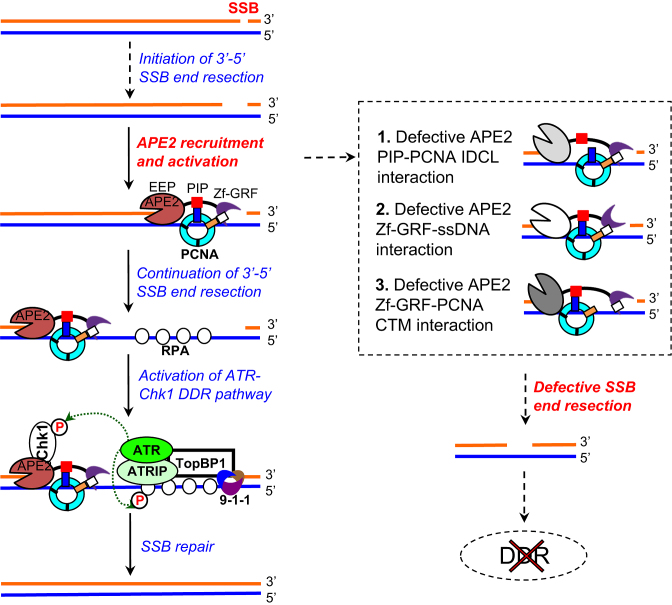
Figure 8.
A working model of the molecular mechanism of APE2-mediated ATR–Chk1 DDR pathway from a defined SSB structure. Please see text for more details.
DISCUSSION
To the best of our knowledge, this is the first report that a defined SSB triggers the ATR–Chk1 DDR pathway in a eukaryotic experimental system. Based on evidence in this study, we propose a working model for the SSB-induced ATR–Chk1 DDR pathway (Figure 8): (a) 3′-5′ SSB end resection is initiated into a small gap (∼1nt–3nt) by a mechanism to be determined; (b) APE2 is recruited by PCNA via its PIP box (24), and activated by APE2 Zf–GRF interaction with ssDNA (32) and PCNA C-terminus for SSB end resection continuation (this study); (c) a longer stretch of ssDNA (∼18–26nt) is generated and bound with RPA, leading to the assembly of ATR-ATRIP, TopBP1, and 9–1–1 complex to activate DDR; (d) activated ATR phosphorylates its substrates including Chk1 and RPA32 and (e) activated ATR DDR pathway is important for SSB repair (this study).
Our SSB signaling system requires only HSS but not the addition of NPE, which is different from previously established reconstitution systems such as ATR DDR pathway activation by 3′-primed ssDNA or defined ICLs using Xenopus HSS and NPE combined systems (55,56). The defined SSB structure is resected by APE2 in the 3′ to 5′ direction to generate a longer stretch of ssDNA, which is in line with the previously established general model for ATR–Chk1 DDR pathway activation (7,8,14,55). To test whether the SSB plasmid triggers ATR activation in NPE, we found that no detectable Chk1 phosphorylation was induced by SSB plasmid in NPE (Supplementary Figure S2B). We reason that some phosphatases of Chk1 in NPE may destabilize the potential Chk1 phosphorylation by activated ATR. To test this possibility, we chose to use tautomycin, which was shown to stabilize Chk1 phosphorylation induced by AT70 in the LSS system (34). Interestingly, with the presence of tautomycin, the SSB plasmid triggered a very robust Chk1 phosphorylation whereas CTL plasmid induced a minimal Chk1 phosphorylation in NPE (Supplementary Figure S2B). Both Chk1 phosphorylation events in NPE were inhibited by the addition of VE-822 (Supplementary Figure S2B). Whereas the minimal Chk1 phosphorylation induced by CTL plasmid in NPE may be due to increased DNA-to-Cytoplasmic ratio (57), the SSB-induced increase of Chk1 phosphorylation suggests that ATR–Chk1 DDR is likely induced by SSB in NPE.
One striking feature of this experimental system is that SSB signaling is replication-independent in the HSS (Figure 2E and F, and Supplementary Figure S2C), which is consistent with the deficiency of DNA replication elongation in the HSS system (37). Because a variety of checkpoint proteins play important roles for DNA replication, our defined SSB signaling system in a cell-free system provides a powerful experimental system for future applications in determining direct roles of candidate checkpoint proteins in DDR pathway but not indirectly through their function in DNA replication. This replication-independent SSB-induced ATR–Chk1 DDR in the HSS system is reminiscent of the DSB-mimicking AT-70 induced replication-independent ATR–Chk1 DDR in the LSS system (34,51). As we estimate that the ssDNA gap after SSB end resection is ∼18nt–26nt (Figure 6B), the ssDNA gap is likely to be filled by DNA polymerases for SSB repair. Future work is needed to test whether such repair DNA synthesis is important for ATR activation.
The two modes of APE2–PCNA interaction are intriguing. APE2 interacts with PCNA’s IDCL motif via its PIP box and associates with ssDNA via its Zf–GRF motif (24,30–32). Importantly, we found that APE2 Zf–GRF also interacts with PCNA, mainly through PCNA’s CTM region. Therefore, we propose two modes of APE2–PCNA interaction: APE2 PIP box-PCNA IDCL interaction and APE2 Zf–GRF–PCNA CTM interaction, which are designed as Mode I and Mode II interaction, respectively (Figure 5A). Although it is not explicitly determined as of now how the two modes of APE2–PCNA interaction are selected and/or transitioned dynamically, a previous study in yeast showed that APE2 interacts with PCNA IDCL with the absence of DNA and switches to PCNA C-terminal tail upon entering a 3′ primer-template junction (31). Our findings identified that APE2 Zf–GRF interacts with PCNA CTM even in the absence of DNA (Figure 5). Furthermore, it is our understanding that the two modes of APE2–PCNA interaction are neither required for nor mutually exclusive to each other. We interpret the observations that the Mode I interaction plays a major role in APE2 recruitment to SSB sites whereas the Mode II interaction plays a critical role in APE2 activation in the HSS system. Nonetheless, the biological significance of Mode II of APE2–PCNA interaction is evidenced by deficiency of SSB end resection and SSB signaling by the mutant C470A APE2 in the HSS system (Figure 6A). Why is the Mode II of APE2–PCNA interaction needed for APE2 activation in SSB signaling? We speculate that the Mode II of APE2–PCNA interaction is needed to overcome the inhibition of APE2 by a previously unidentified negative regulator for SSB end resection in the HSS system. The Model I of APE2–PCNA interaction may bring APE2 to PCNA-bound DNA even under normal conditions. However, APE2 is not activated until the Mode II interaction makes the appropriate confirmation change of the APE2–PCNA–DNA complex to stimulate APE2’s exonuclease activity. Notably, the ssDNA interaction via APE2’s Zf–GRF is also critical for APE2 activation (Figure 8) (32). These three distinct mechanisms of APE2 recruitment and activation are essential to ensure that SSB end resection only takes place at the right SSB sites for genome stability.
In addition, both IDCL and CTM regions within PCNA are important for PCNA-stimulated 3′-5′ exonuclease activity of APE2 in vitro (Figure 7A). To examine the interaction of PCNA and APE2 to gapped DNA structure, we established an in vitro protein-DNA binding approach with biotin-gapped dsDNA coupled to streptavidin dynabeads (Supplementary Figure S6A). Notably, GST-APE2, but not GST, was found on Biotin-DNA-coupled beads but not ‘no DNA’ beads, suggesting that APE2 binds to gapped DNA substrate in vitro at least under these experimental conditions. We interpret that the 1–3nt ssDNA gap may be sufficient for APE2 interaction in vitro. Interestingly, the addition of WT PCNA has minimal effect on APE2’s binding to the gapped DNA substrate (Supplementary Figure S6A). Furthermore, LI PCNA, PK PCNA, and LIPK PCNA behave similar to WT PCNA concerning APE2’s binding to the gapped DNA structure (Supplementary Figure S6B). It seems that both modes of PCNA-APE2 interaction are important for APE2 exonuclease activity in vitro. More structural studies are needed in the future to figure out how exactly APE2, PCNA and DNA interact with each other in a dynamic and coordinated fashion.
Previous studies have demonstrated that the homologue residues of E34A and D273A within APE2 from yeast and human cells are exonuclease defective mutants (27,58). We have demonstrated in our previous studies that the E34A APE2 and D273A APE2 failed to rescue the hydrogen peroxide-induced Chk1 phosphorylation in APE2-depleted LSS system (24). To verify the Xenopus E34A APE2 and D273A APE2 are indeed exonuclease deficient mutants, we test their exonuclease activity using FAM-labeled gapped DNA structure as template in vitro. Consistent with previous studies on APE2 in other species, the PCNA-promoted exonuclease activity of Xenopus GST-APE2 was significantly decreased in E34A GST-APE2 and D273A GST-APE2 (Supplementary Figure S5B). Notably, WT, but not E34A or D273A, Myc-APE2 rescued Chk1 phosphorylation in APE2-depleted HSS (‘extract’ panel, Supplementary Figure S5C). Although E34A and D273A Myc-APE2 associated with DNA in a similar fashion as WT Myc-APE2, the recruitment of RPA32 and checkpoint protein complex including ATR, ATRIP, TopBP1, and Rad9 onto SSB plasmid was rescued by WT, but not E34A or D273A, Myc-APE2 in APE2-depleted HSS (‘DNA-bound’ panel, Supplementary Figure S5C). These observations suggest that the exonuclease activity of APE2 is indeed important for the SSB-induced ATR activation in the HSS system.
APE2 Zf–GRF interaction with PCNA is a distinct feature compared with ssDNA interaction. There are three types of mutants in APE2 Zf–GRF in terms of PCNA and ssDNA interaction: (I) C470A Zf–GRF and F486A-Y487A Zf–GRF are deficient for PCNA interaction but proficient for ssDNA interaction, (II) R502A APE2 Zf–GRF is deficient for ssDNA interaction but proficient for PCNA interaction (32), and (III) G483A-R484A APE2 Zf–GRF is defective for both PCNA association and ssDNA interaction (Figure 5 and Supplementary Figure S4). These observations suggest that the PCNA association and ssDNA interaction of APE2 Zf–GRF are neither dependent on nor mutually exclusive to each other. The C470 residue of Zf–GRF is localized in the flexible region between Polyproline helix and β1 region (Figure 5A), suggesting that the C470A point mutation may not affect the secondary structure of APE2 Zf–GRF. Because Zf–GRF has been found in more than 100 proteins involved in DNA/RNA metabolism such as NEIL3 and Top3 (32), the Mode II of APE2–PCNA interaction may be applicable to future structure-function studies of other Zf–GRF-containing proteins.
SSB end resection has unique characteristics in comparison to other DNA end processing pathways such as DSB end resection. In this study, we present our substantial data to show that a site-specific SSB structure triggers an ATR–Chk1 DDR pathway via SSB end resection in a eukaryotic cell-free system. One distinct feature of SSB end resection is the critical role of APE2’s 3′-5′ exonuclease activity (Figure 6D). Although a nicked dsDNA structure is resected by recombinant APE2 in vitro (27), the FAM-SSB structure is still catalyzed into Type II resected products by other DNA end processing enzymes in APE2-depleted HSS (Figure 6D). There are two possible explanations: APE2 plays an important role in both the initiation and continuation of 3′-5′ SSB end resection in the HSS system, and another unknown 3′-5′ exonuclease may compensate the initiation of 3′-5′ SSB end resection in APE2-depleted HSS. Alternatively, a previously unidentified 3′-5′ exonuclease is needed to initiate SSB end resection followed by continuation of 3′-5′ SSB end resection by APE2. Of note, no apparent Type II resected products were observed in our time-course experiment (Figure 6B). We speculate that the initiation phase of 3′-5′ SSB end resection takes time to complete, and that end resection continuation is much quicker as long as a short ssDNA gap is generated. More experiments are needed to test these different possibilities in the future. DSB end resection in the 5′-3′ direction coupling DSB repair and DDR pathways has been investigated extensively (18). Exo1-meidated 5′-3′ DSB end resection has also been implicated in DSB repair, nucleotide excision repair (NER) and mismatch repair (MMR) pathways (59–61). Whereas Mre11 participates in 3′-5′ end resection of protein-DNA adducts, a critical question remains unanswered whether the Mre11’s 3′-5′ end resection plays a critical role for DDR pathway activation (62).
In addition, previous studies demonstrate that ATR and ATRIP preferentially bind to approximately 70nt to 80nt ssDNA coated with RPA in in vitro binding assays (10). Consistent with this, a gapped plasmid with 68nt ssDNA is sufficient to trigger an ATR–Chk1 DDR pathway in a DNA-PKcs-dependent manner in human cell-free extracts (63). To test whether ATR and ATRIP are recruited to this short stretch of ssDNA (18nt-26nt), we found that as short as 20nt ssDNA is sufficient for binding of RPA, ATR and ATRIP in the HSS system (Figure 6C). This observation is consistent with the preferential recruitment of ATR and ATRP to SSB sites in the HSS system shown in Figure 6A. Future studies will focus on how the SSB end resection is terminated to promote SSB repair.
From COSMIC analysis (cancer.sanger.ac.uk/cancergenome/projects/cosmic) of 45 cancer patients with somatic mutations in APE2, 33 missense point mutations were found in APE2, out of which 21 mutant residues of human APE2 are identical to Xenopus APE2 in homologue analysis. These 21 substitution missense point mutants in human APE2 are converted into Xenopus APE2: G10E, T38S, V49I, G51S, R62H, A69S, A79S, E83G, L110R, E152K, R159C, R208C, R244C, R264H, H300Q, A314T, E343K, A366V, G456E, E468G and R484H. We found that 15 residues are in the N-terminal EEP domain and two mutant residues (E468 and R484) are in the Zf–GRF domain. In particular, the R484H mutant within APE2 Zf–GRF may be deficient for PCNA interaction and ssDNA interaction. In addition, one nonsense substitution in human APE2 (c.1342G>T, p.E448*) was confirmed as a somatic mutation in a lung carcinoma patient (TCGA-75-6211-01), leading to a mutant APE2 protein that lacks the whole Zf–GRF domain. These somatic mutations in cancer patients suggest that the Zf–GRF domain of APE2 may have biological significance via its critical function in SSB signaling. Dysfunctions in DNA repair and DDR signaling pathways are implicated in cancer development (6). Importantly, a variety of DNA repair and DDR proteins including ATR and Chk1 have become therapeutic targets and are currently being tested in the laboratory and clinical studies (64). A better understanding of DDR pathway activation in response to SSBs has implications in new avenues of cancer treatment. Findings from our experiments will impact future translational studies such as anti-cancer therapies via the modulation of novel role of APE2 in SSB signaling using mammalian cell lines or mice models. In addition, a recent report demonstrates that sea squirt APE2 is more likely involved in oxidative stress response than in editing function via its 3′-5′ exonuclease activity, suggesting the conserved function of APE2 during evolution (65).
Overall, our evidence using defined SSB end resection and SSB signaling in Xenopus provides novel insights into SSB-induced DDR pathway by APE2 in a eukaryotic cell-free system.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Scott Williams and associates for insightful discussions on this study. We thank Drs Matthew Michael, Karlene Cimprich, Howard Lindsay, Yusaku Nakabeppu, Daisuke Tsuchimoto and Yoshiaki Azuma for reagents. We thank Dr Didier Dréau for assistance on FACS analysis of cells. We also thank Drs Yvette Huet and Chandra Williams as well as Hernando Gordils and Alvaro Perez for maintaining the health of our frogs.
Author contributions: S.Y. conceived the SSB signaling technology. Y.L., L.B. and S.Y. designed and performed the experiments. S.C., M.A.H., B.D., M.M. and J.R. performed the experiments. S.Y. supervised the project and wrote the manuscript with input from all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health [R15GM101571, R15GM114713 to S.Y.]; Duke Energy Endowment Special Initiatives Fund and Faculty Research Grant by University of North Carolina at Charlotte [to S.Y.]; National Science Foundation [REU 1359271 to J.R.]. Funding for open access charge: Subvention Fund (OAAPSF) at UNC Charlotte and the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008; 9:619–631. [DOI] [PubMed] [Google Scholar]
- 2. Yan S., Sorrell M., Berman Z.. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol. Life Sci. 2014; 71:3951–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andres S.N., Schellenberg M.J., Wallace B.D., Tumbale P., Williams R.S.. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. 2015; 56:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee D.H., Liu Y., Lee H.W., Xia B., Brice A.R., Park S.H., Balduf H., Dominy B.N., Cao W.. A structural determinant in the uracil DNA glycosylase superfamily for the removal of uracil from adenine/uracil base pairs. Nucleic Acids Res. 2015; 43:1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davis L., Maizels N.. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E924–E932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cimprich K.A., Cortez D.. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell. Biol. 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith K.D., Fu M.A., Brown E.J.. Tim-Tipin dysfunction creates an indispensible reliance on the ATR–Chk1 pathway for continued DNA synthesis. J. Cell Biol. 2009; 187:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 11. Yan S., Michael W.M.. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 2009; 184:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J., Gong Z., Chen J.. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J. Cell Biol. 2011; 193:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Acevedo J., Yan S., Michael W.M.. Direct binding to replication protein A (RPA)-coated single-stranded DNA allows recruitment of the ATR activator TopBP1 to sites of DNA damage. J. Biol. Chem. 2016; 291:13124–13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. . ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 16. Guo Z., Kumagai A., Wang S.X., Dunphy W.G.. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000; 14:2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bakkenist C.J., Kastan M.B.. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003; 421:499–506. [DOI] [PubMed] [Google Scholar]
- 18. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 19. You Z., Bailis J.M.. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010; 20:402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T.. ATM activation by oxidative stress. Science. 2010; 330:517–521. [DOI] [PubMed] [Google Scholar]
- 21. Khoronenkova S.V., Dianov G.L.. ATM prevents DSB formation by coordinating SSB repair and cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3997–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson C., Jasin M.. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000; 405:697–700. [DOI] [PubMed] [Google Scholar]
- 23. Rudin N., Haber J.E.. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 1988; 8:3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willis J., Patel Y., Lentz B.L., Yan S.. APE2 is required for ATR–Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okano S., Lan L., Caldecott K.W., Mori T., Yasui A.. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003; 23:3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tell G., Quadrifoglio F., Tiribelli C., Kelley M.R.. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Sign. 2009; 11:601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burkovics P., Szukacsov V., Unk I., Haracska L.. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006; 34:2508–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guikema J.E., Gerstein R.M., Linehan E.K., Cloherty E.K., Evan-Browning E., Tsuchimoto D., Nakabeppu Y., Schrader C.E.. Apurinic/apyrimidinic endonuclease 2 is necessary for normal B cell development and recovery of lymphoid progenitors after chemotherapeutic challenge. J. Immunol. 2011; 186:1943–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moldovan G.L., Pfander B., Jentsch S.. PCNA, the maestro of the replication fork. Cell. 2007; 129:665–679. [DOI] [PubMed] [Google Scholar]
- 30. Burkovics P., Hajdu I., Szukacsov V., Unk I., Haracska L.. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 2009; 37:4247–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unk I., Haracska L., Gomes X.V., Burgers P.M.J., Prakash L., Prakash S.. Stimulation of 3 ‘→ 5 ’ exonuclease and 3′-phosphodiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol. Cell. Biol. 2002; 22:6480–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wallace B.D., Berman Z., Mueller G.A., Lin Y., Chang T., Andres S.N., Wojtaszek J.L., DeRose E.F., Appel C.D., London R.E. et al. . APE2 Zf–GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lupardus P.J., Byun T., Yee M.C., Hekmat-Nejad M., Cimprich K.A.. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 2002; 16:2327–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumagai A., Dunphy W.G.. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000; 6:839–849. [DOI] [PubMed] [Google Scholar]
- 35. Lebofsky R., Takahashi T., Walter J.C.. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 2009; 521:229–252. [DOI] [PubMed] [Google Scholar]
- 36. Cupello S., Richardson C., Yan S.. Cell-free Xenopus egg extracts for studying DNA damage response pathways. Int. J. Dev. Biol. 2016; 60:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walter J., Sun L., Newport J.. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998; 1:519–529. [DOI] [PubMed] [Google Scholar]
- 38. Philpott A., Yew P.R.. The Xenopus cell cycle: an overview. Mol. Biotechnol. 2008; 39:9–19. [DOI] [PubMed] [Google Scholar]
- 39. Willis J., Destephanis D., Patel Y., Gowda V., Yan S.. Study of the DNA damage checkpoint using Xenopus egg extracts. J. Vis. Exp. 2012; e4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones R.E., Chapman J.R., Puligilla C., Murray J.M., Car A.M., Ford C.C., Lindsay H.D.. XRad17 is required for the activation of XChk1 but not XCds1 during checkpoint signaling in Xenopus. Mol. Biol. Cell. 2003; 14:3898–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lupardus P.J., Cimprich K.A.. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol. Biol. Cell. 2006; 17:1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bai L., Michael W.M., Yan S.. Importin beta-dependent nuclear import of TopBP1 in ATR–Chk1 checkpoint in Xenopus egg extracts. Cell Signal. 2014; 26:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S.P., Dasso M., Azuma Y.. PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J Biol Chem. 2010; 285:14415–14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y.. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001; 29:2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson D.M., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J. Mol. Biol. 2003; 330:1027–1037. [DOI] [PubMed] [Google Scholar]
- 46. Kumagai A., Lee J., Yoo H.Y., Dunphy W.G.. TopBP1 activates the ATR-ATRIP complex. Cell. 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- 47. Duursma A.M., Driscoll R., Elias J.E., Cimprich K.A.. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol. Cell. 2013; 50:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Post S.M., Tomkinson A.E., Lee E.Y.. The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase epsilon. Nucleic Acids Res. 2003; 31:5568–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X., Zou L., Lu T., Bao S., Hurov K.E., Hittelman W.N., Elledge S.J., Li L.. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 2006; 23:331–341. [DOI] [PubMed] [Google Scholar]
- 50. Stokes M.P., Van Hatten R., Lindsay H.D., Michael W.M.. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 2002; 158:863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yan S., Lindsay H.D., Michael W.M.. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 2006; 173:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gamper A.M., Rofougaran R., Watkins S.C., Greenberger J.S., Beumer J.H., Bakkenist C.J.. ATR kinase activation in G1 phase facilitates the repair of ionizing radiation-induced DNA damage. Nucleic Acids Res. 2013; 41:10334–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Whitehouse C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W.. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001; 104:107–117. [DOI] [PubMed] [Google Scholar]
- 54. Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D.. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol. Cell. 2015; 60:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. MacDougall C.A., Byun T.S., Van C., Yee M.C., Cimprich K.A.. The structural determinants of checkpoint activation. Genes Dev. 2007; 21:898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ben-Yehoyada M., Wang L.C., Kozekov I.D., Rizzo C.J., Gottesman M.E., Gautier J.. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell. 2009; 35:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Conn C.W., Lewellyn A.L., Maller J.L.. The DNA damage checkpoint in embryonic cell cycles is dependent on the DNA-to-cytoplasmic ratio. Dev. Cell. 2004; 7:275–281. [DOI] [PubMed] [Google Scholar]
- 58. Unk I., Haracska L., Prakash S., Prakash L.. 3′-phosphodiesterase and 3′→5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 2001; 21:1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lindsey-Boltz L.A., Kemp M.G., Reardon J.T., Derocco V., Iyer R.R., Modrich P., Sancar A.. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 2014; 289:5074–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garcia V., Phelps S.E., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen X., Paudyal S.C., Chin R.I., You Z.. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013; 41:9325–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deshpande R.A., Lee J.H., Arora S., Paull T.T.. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell. 2016; 64:593–606. [DOI] [PubMed] [Google Scholar]
- 63. Vidal-Eychenie S., Decaillet C., Basbous J., Constantinou A.. DNA structure-specific priming of ATR activation by DNA-PKcs. J. Cell Biol. 2013; 202:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Curtin N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer. 2012; 12:801–817. [DOI] [PubMed] [Google Scholar]
- 65. Funakoshi M., Nambara D., Hayashi Y., Zhang-Akiyama Q.-M.. CiAPEX2 and CiP0, candidates of AP endonucleases in Ciona intestinalis, have 3′-5′ exonuclease activity and contribute to protection against oxidative stress. Genes Environ. 2017; 39:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.