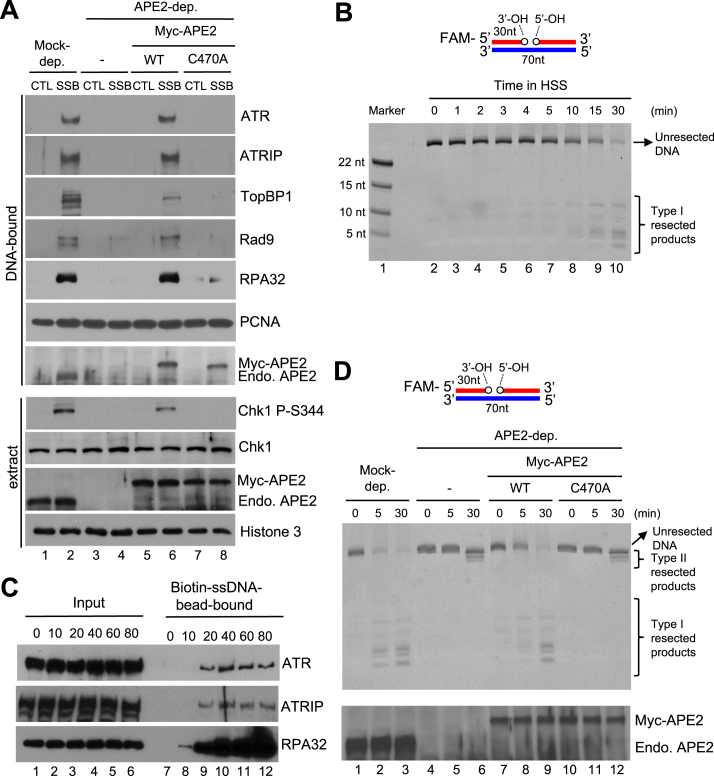
Figure 6.
APE2 Zf–GRF–PCNA interaction promotes SSB end resection, the assembly of a checkpoint protein complex onto SSB plasmid, and Chk1 phosphorylation in the HSS system. (A) CTL or SSB plasmid was added to mock- or APE2-depleted HSS, which was supplemented with WT or C470A Myc-APE2. DNA-bound fractions and total extract samples were examined via immunoblotting analysis as indicated. ‘Endo. APE2’ represents endogenous APE2. (B) FAM-labeled dsDNA with a site specific SSB (designed as FAM-SSB) was added to HSS for different time as indicated. Then samples were examined via TBE-Urea gel and visualized via Typhoon imager. ‘Marker’ represents four FAM-labeled different-length ssDNA. (C) The length dependence of ssDNA for the recruitment of ATR-ATRIP complex and RPA to ssDNA in the HSS. Streptavidin Dynabeads coupled with different length of Biotin-coupled ssDNA (i.e., 0, 10, 20, 40, 60 or 80nt) were added to HSS. After incubation, the Biotin-ssDNA bead-bound fractions were isolated from HSS. The Input and bead-bound fractions were examined via immunoblotting analysis. (D) The FAM-SSB substrate was added to mock- or APE2-depleted HSS, which was supplemented with WT or C470A Myc-APE2. DNA structures were examined via TBE-Urea gel and visualized via Typhoon imager (top). Samples were also analyzed via immunoblotting analysis as indicated. ‘Endo. APE2’ represents endogenous APE2 (bottom).