Figure 1.
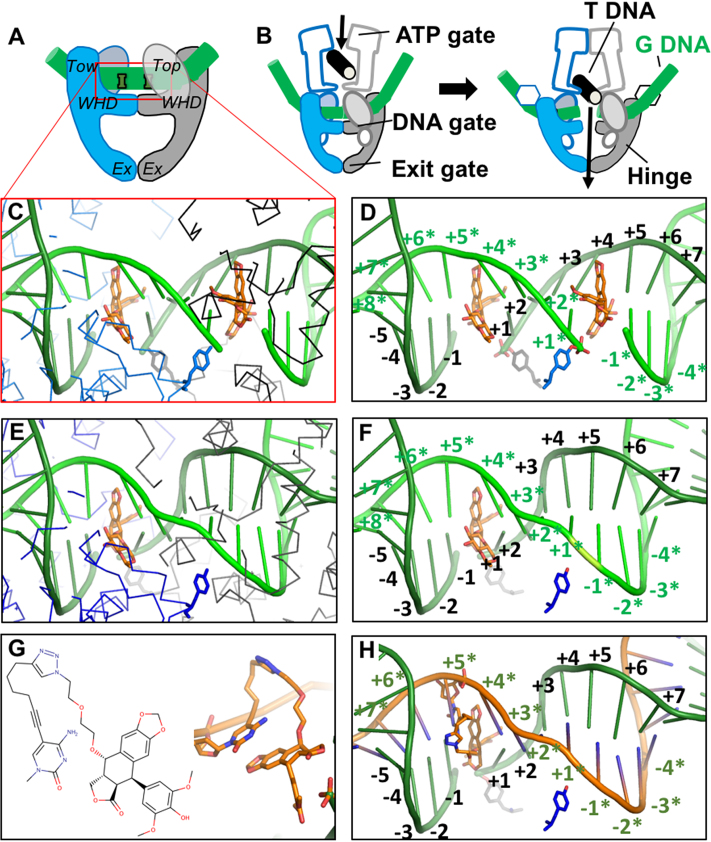
Structure-guided design of an OTI. (A) Schematic illustrating domains of type II topoisomerases used to determine the crystal structure of human topoisomerase IIβ covalently attached to DNA (green) in the presence of etoposide (orange). Domains pictured are TOPRIM (Top), winged helix domain (WHD), tower domain (Tow), and exit gate domain (Ex). (B) Schematic of topoisomerase II function. Protein protomer subunits are shown in blue and gray. T DNA, transport double helix (black); G DNA, gate double helix (green). (C, D) Detail from the crystal structure of a topoisomerase IIβ cleavage complex with two bound etoposide molecules (orange) stabilizing a double-stranded DNA (green) break; PDB code 3QX3. For clarity, in panel C, only the Cα trace of the protein subunits (blue and black lines) and catalytic tyrosines (blue and gray sticks) are shown. In panel D, only the catalytic tyrosine residues that cleave the DNA are shown. The conventional numbering scheme used for DNA cleavage complexes formed by type II topoisomerases is shown. The enzyme cleaves between the -1 and the +1 on each strand. The numbering on the two strands in the double helix is differentiated by the presence or absence of asterisks. The catalytic tyrosine residues are covalently attached to the DNA at the +1 positions. (E, F) Model of a cleavage complex with one bound etoposide molecule stabilizing a single-stranded DNA break. The cleaved DNA strand is indicated by asterisks. The protein subunits shown are the same as those in C and D. (G) Chemical (left) and modeled (right) structure of the etoposide core (DEPT) linked to the pyrimidine base. (H) OTI28 (orange strand) modeled with the modified cytosine base in the +5* position, stabilizing DNA scission at the 23–24 site (–1 to +1) on the cleaved target strand (green). Structural figures were drawn with Pymol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).