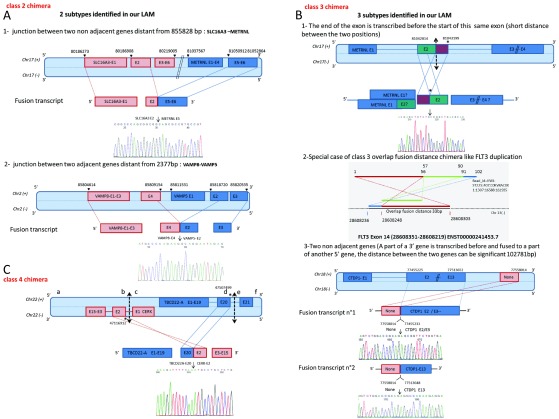
Figure 4. Different classes of validated chRNA.
The position and orientation of the genes on the chromosomes have been schematically arranged. The start and end positions of the exons have been indicated. The representations of the chRNA junction supported by the Sanger sequencing results are also shown. ( A) Two subtypes illustrating Class 2 chimera junctions. The first subtype 1 is illustrated by the junction of two non-adjacent genes SLC16A3 and METRNL, separated by 855828 bp. The second subtype 2 is illustrated by the junction between adjacent genes VAMP8 and VAMP5, separated by 2377 bp. ( B) Three subtypes illustrating Class 3 chimera junction. They all involve the inversion of exon order, first two concern an inversion within a same exon of a gene (METRNL and FLT3 cases), and the third subtype involves an inversion of a transcribed sequence, separated by about 103 000 bp (NONE-CTDP1). ( C) A Class 4 chimeric construction joining an exon from (+) strand gene TBCD22A with an exon from (-) strand gene CERK, arising from the same chromosome 22.