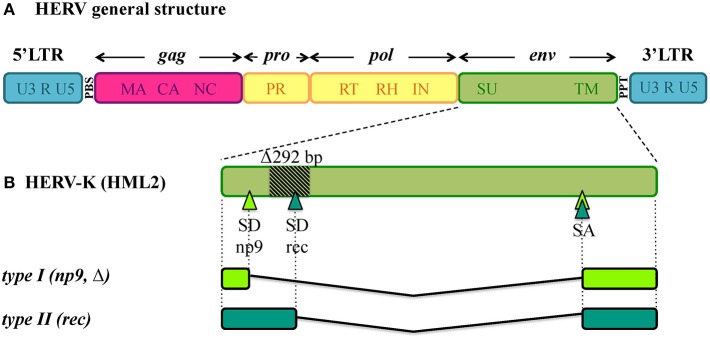
Figure 1.
General structure of HERV DNA sequences. (A) Typical structure of HERV proviruses. The general structure of a full-length HERV proviral sequence is depicted: two Long Terminal Repeats (LTRs) flank gag, pro, pol, and env genes. The viral genes and the correspondent protein products are indicated: gag matrix (MA), capsid (CA) and nucleocapsid (NC); pro-pol protease (PR) – reverse transcriptase (RT), ribonuclease H (RH) and integrase (IN); env surface (SU) and transmembrane (TM). The primer binding site (PBS) and polypurine tract (PPT) are located between 5′LTR and gag and between env and 3′LTR, respectively. It is worth noting that the action of cellular editing systems and the persistence within the host genome led to the accumulation of substitutions, deletion and insertions that modified the structure of the majority of HERV proviruses, leading to coding-defective sequences and solitary LTRs formation. (B) HERV-K(HML2) accessory genes. Differently from other HERV groups, the HERV-K(HML2) env gene presents alternative splicing sites, leading to the presence of a full length transcript with structural significance plus two types of accessory variants, named rec and np9, that subdivide the HML2 sequences into two types. Type I HML2 elements present a characteristic 292-bp env deletion, leading to the use of an upstream splice donor and the subsequent production of a shorter protein, named Np9; while type II HML2 sequences retained such portion and use the downstream splice donor site to encode for a longer protein, named Rec.