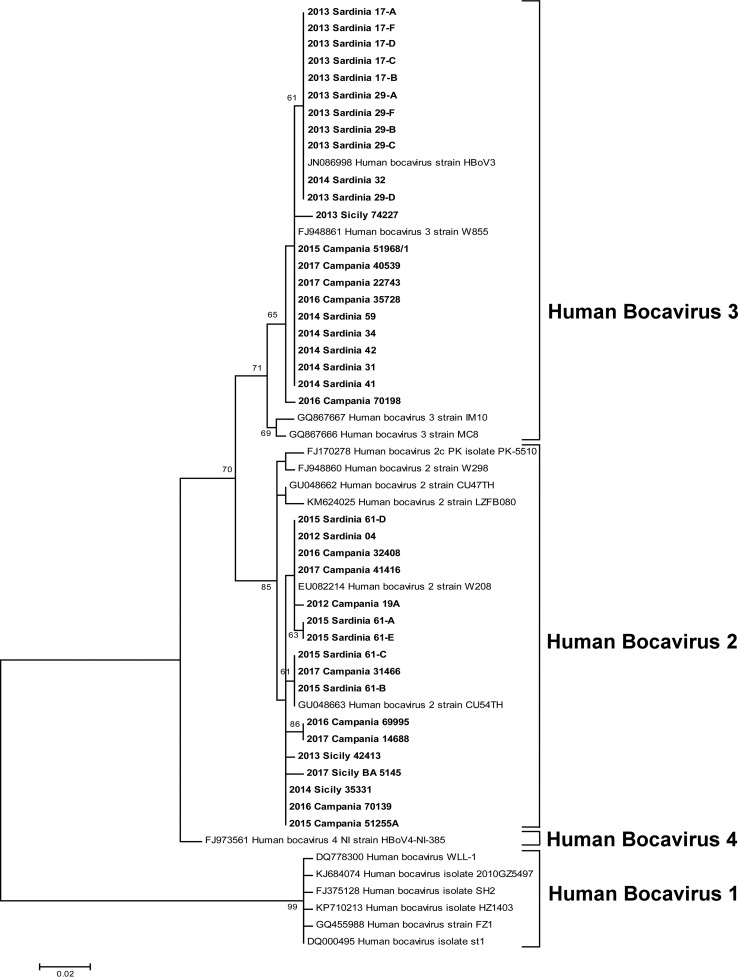
FIG 1.
Phylogenetic tree constructed with the partial HBoV VP1/2 sequences. The tree includes 39 study sequences and 18 genomic sequences available from GenBank, representing species 1 (accession numbers DQ778300, KJ684074, FJ375128, GQ455988, KP710213, and DQ000495), species 2 (accession numbers EU082214, GU048663, FJ170278, FJ948860, GU048662, and KM624025), species 3 (GQ867667, GQ867666, FJ948861, and JN086998), and species 4 (FJ973561, KC461233, and KM624027). Evolutionary analyses were conducted with MEGA7. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura 3-parameter model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Only bootstrap values of >60% are represented. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. There were a total of 300 positions in the final data set.