Figure 6.
Analysis of Public RNA-Seq Datasets Substantiates Identified MO Side Effects
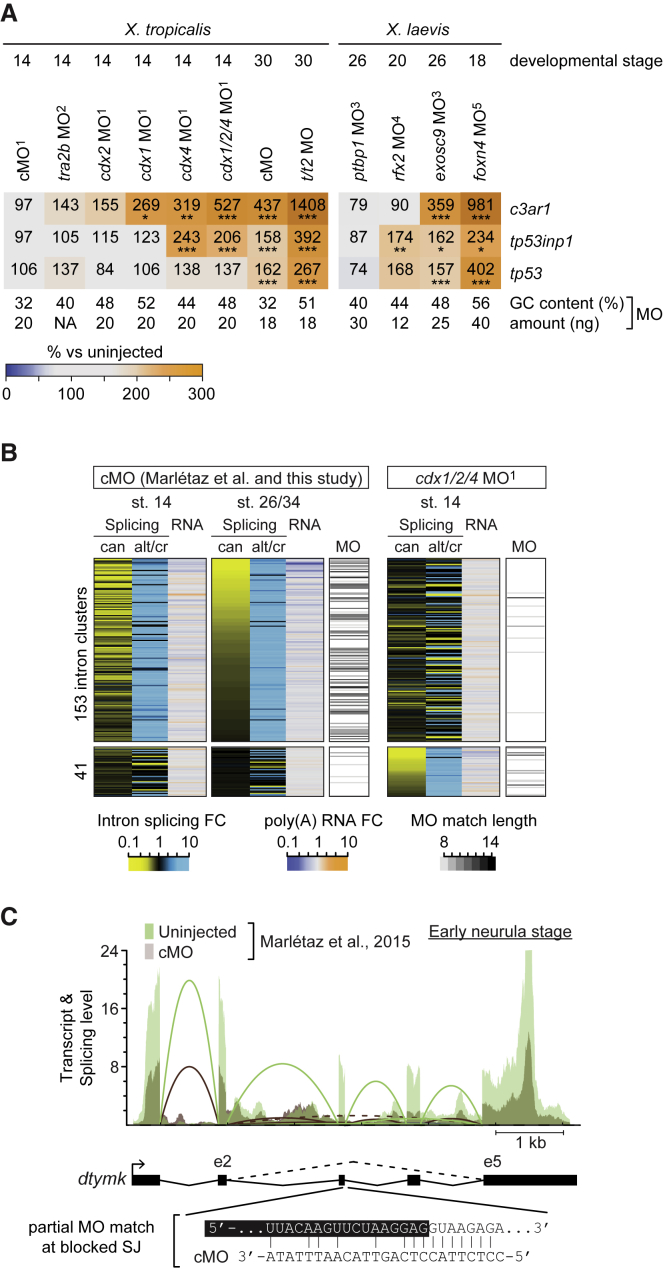
Superscripts refer to the published datasets used in this study: 1Marlétaz et al., 2015; 2Dichmann et al., 2015; 3Noiret et al., 2016; 4Chung et al., 2014; and 5Campbell et al., 2016.
(A) Heatmap of MO-induced transcriptional mis-regulation (%, percentage compared with uninjected embryos) of c3ar1 (c3ar1.L), tp53inp1 (tp53inp1.L), and tp53 (tp53.L) in X. tropicalis and X. laevis (gene names in brackets) embryos at indicated developmental stages. Asterisks indicate statistical significance: ∗FDR ≤10%; ∗∗FDR ≤1%; and ∗∗∗FDR ≤0.1%. The (average) GC content and dosage of MO(s) are listed below the heatmap.
(B) Seriated heatmap of splice/transcript levels and MO match length at blocked splice junctions for MO-injected embryos compared with their uninjected controls. Selected intron clusters represent coupled splicing events that were inversely mis-regulated in embryos injected with cMO (153 intron clusters at tailbud stage) or the cdx1/2/4 MO mix (41 intron clusters at neurula stage): inhibition of canonical (can) splicing caused alternative or cryptic (alt/cr) splice sites to be used more frequently. Intron cluster-specific heatmap rows were sorted by the mis-regulation of canonical splicing in morphants.
(C) Superimposed Sashimi plot of transcript dtymk whose splicing was affected by cMO at early neurula stage (data from Marlétaz et al., 2015). Canonical and alternative (between exons 2 and 5) splicing are shown with solid and dashed lines, respectively. The blocked donor splice site of intron 3 contains 10 consecutive bases perfectly complementary to sequence of the cMO.