Figure 1.
TALEN-Mediated Gene Targeting in Arg1Δ Mouse iPSCs
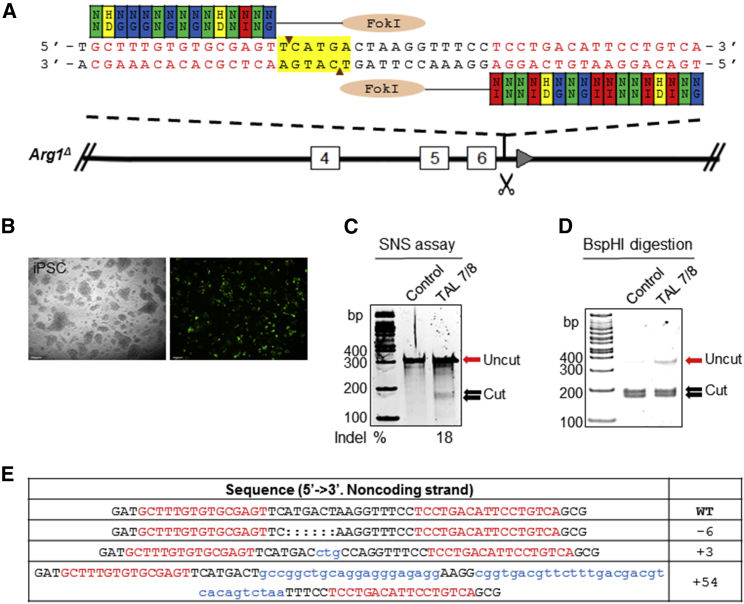
(A) Schematic diagram showing the site of Arg1 gene modification using TALEN set 7/8. The TALEN 7/8 pair was designed to target intron 6 of Arg1. Each TALEN arm consists of a DNA-binding domain with repeat variable di-residues (RVDs) corresponding to DNA binding sequence proceeded by a 5′ T nucleotide and a 17-bp spacer region containing a BspHI recognition site (highlighted in yellow) to assay activity. TALE repeat domains are colored to indicate the identity of the RVD. Site-specific double-stranded breaks (DSBs) are generated upon dimerization of fused FokI endonucleases. (B) Electroporation to deliver 7 μg of each TALEN and pMax-GFP into cells to assess transfection efficiency. Images were acquired 24 hr post-electroporation. Scale bars, 95 μm. (C) Surveyor nuclease assay for detection of NHEJ-induced indels resulting from DSBs. The cleavage products were shown as extra bands (between 162 and 186 bp) indicated by the arrows. Mutation frequencies (indels %) were calculated by measuring the band intensities. (D) BspHI digestion results. The cut products were shown as extra bands (169 bp + 183 bp). (E) Sequencing data from PCR amplicons of TALEN-modified genomic DNA showing a few examples of NHEJ-mediated indel mutations at the desired location. The wild-type sequence is shown above with the TALEN-binding sites in red. Deleted bases are indicated by colons, and inserted bases are shown by lowercase letters in blue. The net change in length caused by each indel mutation is to the right of each sequence.