Figure 2.
Footprint-free TALEN-Mediated Correction of Arg1Δ Mouse iPSCs
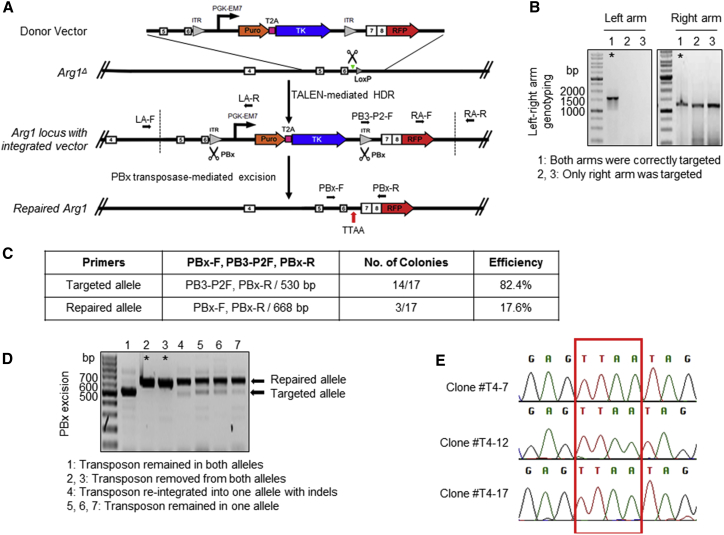
(A) Schematic of strategy used for TALEN-mediated reincorporation of Arg1 exons 7 and 8 via homology-directed repair in combination with piggyBac transposon methodology. Green triangle denotes the target site of TALEN 7/8 in intron 6 of Arg1. Black arrows indicate primers for PCR-based screening to confirm the selected clones. The remnant LoxP left from the initial Cre-excision of exons 7 and 8 would be removed upon targeting vector integration. The characteristic “footprint” TTAA sequence at the site of transposon excision is shown. PBx, piggyBac transposase; T2A, viral sequence for ribosomal skipping; Puro, puromycin; ITR, inverted terminal repeat; TK, thymidine kinase. (B) Representative gel images showing integration-specific PCR of puromycin-resistant single-cell clones derived from Arg1Δ iPSC after TALEN treatment. Each homology arm was amplified independently by PCR. Amplicon sizes of the left and right arm were 1,530 and 1,233 bp, respectively. The clones indicated by an asterisk are correctly targeted with integrated selection cassette and corrective sequence in the desired position at the Arg1 locus. (C) PCR-based excision screening. A table shows primer combination to uniquely identify different alleles after piggyBac excision and negative selection by ganciclovir. (D) A representative gel of different banding patterns and corresponding genotypes. Amplicon sizes of the repaired and targeted alleles were 668 bp and 530 bp, respectively. PBx-excised clones are indicated by asterisks. (E) Footprint sequencing analysis of transposon-free repair clones. TTAA target sites are boxed.