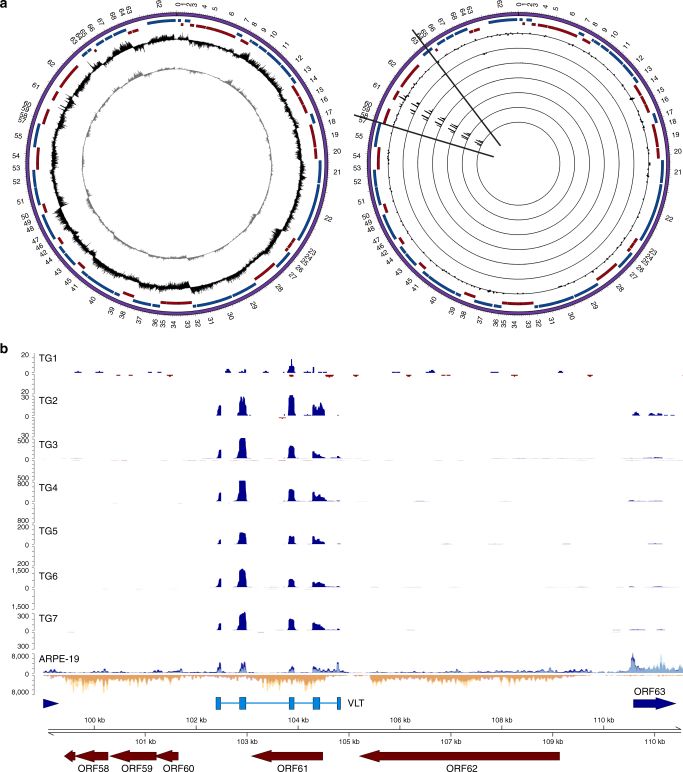
Fig. 2.
VZV transcriptome profile during lytic and latent infection. Strand-specific mRNA-seq of lytically VZV-infected ARPE-19 cells and seven latently VZV-infected human trigeminal ganglia (TG) (Supplementary Table 1). a Circos plots of the VZV genome [purple band; sense and antisense open reading frames (ORFs) indicated as blue and red blocks, respectively], with internal tracks revealing the lytic (left) and latent (right) transcriptomes using unenriched (grey track, left panel) and VZV-enriched (black tracks, left and right panels) libraries. Right panel: latent VZV transcriptome of seven TG, with each track depicting a single specimen. Peaks facing outward from the centre indicate reads mapping to the sense strand, while peaks facing inward originate from the antisense strand. The y axis is scaled to maximum read depth per library in all cases. b Linear representation of the varicella latency-associated transcript (VLT) genomic region (black lines in a), with blue and yellow tracks depicting VZV-enriched mRNA-Seq reads originating from the sense and antisense strands, respectively. Unenriched mRNA-Seq tracks for ARPE-19 cells, and TGs 1 and 2, are superimposed and shown in light blue (sense) or yellow (antisense), with overlapping regions in medium-blue and orange, respectively. No VZV-mapping reads were obtained from unenriched sequence data sets generated from TGs 1 and 2. VZV genome coordinates are shown the VZV reference strain Dumas (NC_001348.1); blue and red arrows indicate previously described VZV ORFs, and light blue boxes indicate the five VLT exons. Paired-end read data sets were generated with read lengths of 2 × 34 bp (ARPE-19) or 2 × 76 bp (TG1 and TG2) or 2 × 151 bp (TG3–TG7)