Figure 1.
SLIM-ChIP Is Robust to MNase and Input Levels
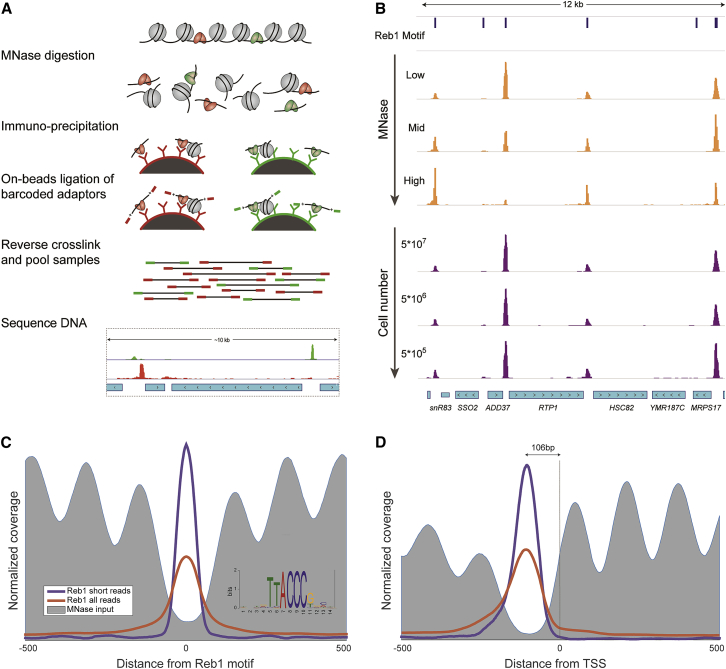
(A) Outline of the SLIM-ChIP method. MNase-digested cross-linked chromatin is barcoded on beads allowing for multiplexing and efficient capturing of small transcription factor-bound DNA fragments.
(B) Genome browser view of Reb1 binding across range of MNase digestion (1X,9X,27X) and input material levels. The top track (Reb1 motif) marks consensus Reb1-binding motifs.
(C and D) Reb1-binding peaks are depleted for nucleosomes (C) and positioned at a typical distance from the TSS (D). The MNase input track was adapted from Weiner et al. (2015). Fragments of less than 80 bp were considered short reads. The Reb1 logo was generated for peaks called using short-reads data.
See also Figures S1 and S2.