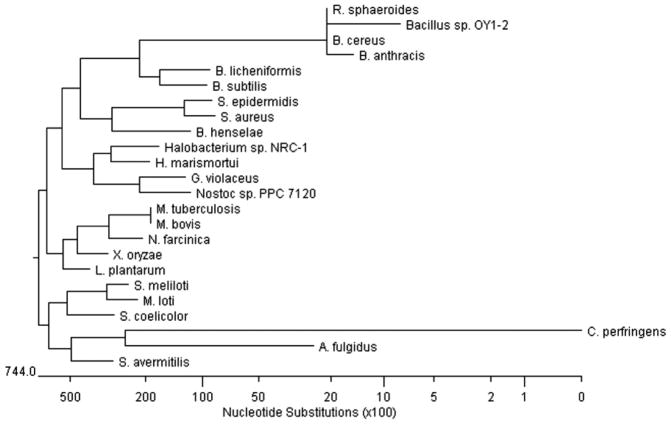
Fig. 4.
Unrooted phylogenetic tree of NADPH-flavin azoreductases. Based on TBALSTN results, protein sequences showing similarities were aligned using Jutun Hein method. The unrooted tree was generated using MegAlign of DNASTAR software package. Genbank accession numbers are as follows: Archaeoglobus fulgidus, AE000972; Bacillus anthracis, AE017225; Bacillus cereus, AE017005; Bacillus licheniformis, CP000002; Bacillus subtilis, AB071366; Bacillus sp. OY1-2; Bartonella henselae, BX897699; Clostridium perfringens, BA000016; Gloeobacter violaceus, BA000045; Haloarcula marismortui AY596297; Halobacterium sp. NRC-1, AE005072; Lactobacillus plantarum, AL935261; Mesorhizobium loti, BA000045; Mycobacterium bovis, BX248344; Mycobacterium tuberculosis, AE000516; Nocardia farcinica, AP006618; Nostoc sp. PPC 7120, BA000019; Rhodobacter sphaeroides, AY150311; Staphylococcus aureus, AY545994; Staphylococcus epidermidis, AE016745; Streptomyces avermitilis, AP005049; Streptomyces coelicolor, AL939107; Sinorhizobium meliloti, AL595985; Xanthomonas oryzae AE013598 (Adapted from [61]).