Fig. 7.
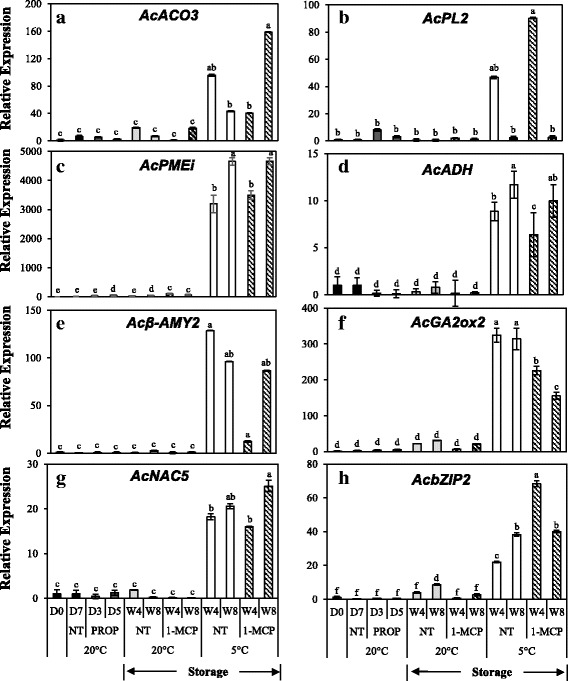
Reverse Transcriptase–Quantitative PCR analysis of selected kiwifruit genes that were exclusively up–regulated by low temperature. Kiwifruit were continuously treated with 5000 μLL− 1 propylene (PROP) at 20 °C, alongside a non–treated group (NT). For storage, kiwifruit were kept at either 5 °C or 20 °C with (1–MCP) or without regular 1–MCP treatment (NT). 1–MCP was applied twice a week at 5 μLL− 1 for ggmxf. Gene–specific primers were designed for (a) AcACO3: ACC OXIDASE 3 (Achn150611); (b) AcPL2: PECTATE LYASE 2 (Achn315151/HQ108112); (c) AcPMEi: PECTIN METHYLESTERASE INHIBITOR (Achn319051/FG458520); (d) AcADH: ALCOHOL DEHYDROGENASE (Achn262421); (e) Acβ–AMY2: β–AMYLASE 2 (Achn212571); (f) AcGA2ox2: GIBBERELLIC ACID OXIDASE 2 (Achn218871); (g) AcNAC5 (Achn169421) and (h) AcbZIP2 (Achn227711). AdACTIN (EF063572) was used as the housekeeping gene and the expression of fruit at harvest (D0) was calibrated as 1. Values are means of three independent biological replicates. Error bars represent SE. Different letters indicate significant differences at p < 0.05. Symbols are D = Day, W = Week, NT = non–treated and PROP = propylene treatment
