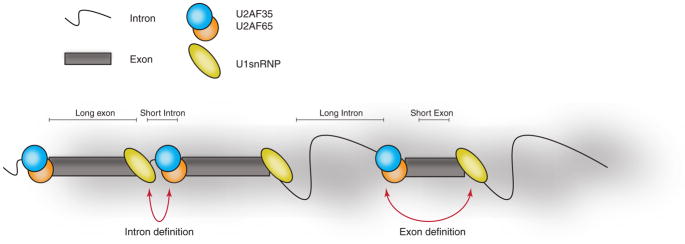
Figure 1. Intron- and exon-definition models.
Schematic representation of the two primary models that drive assembly of splicing factors complexes and define the use of the splice sites in different mRNA architectures. The intron definition model is applicable to lower eukaryotes such as yeasts where the length of the intronic regions is rather small. The exon definition model proposes an explanation for more complex splicing regulation in higher eukaryotes due to the complexity of the genome organization and the presence of large intronic areas that are on average ten times longer than coding exons.