Fig. 7. Inhibition of USP14 impairs histone acetylation.
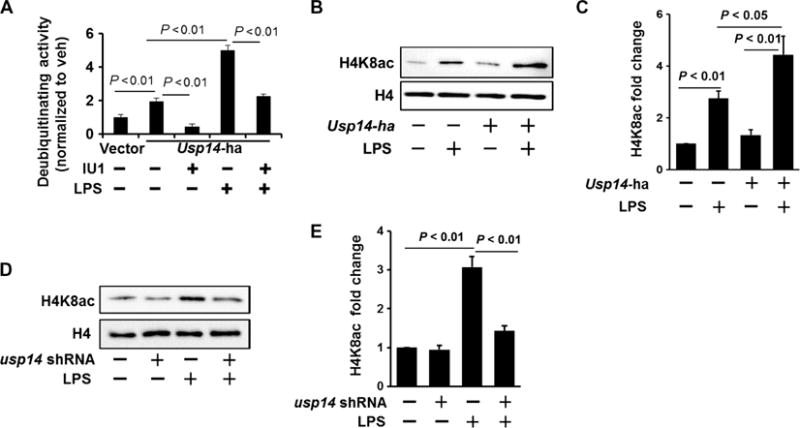
(A) MLE12 cells were transfected with Usp14-ha plasmid. Forty-eight hours later, cells were treated with LPS (10 μg/ml, 1 hour). USP14-HA (hemagglutinin) was immunoprecipitated with an anti-HA antibody, and then the immunoprecipitated complex was incubated with or without IU1. A deubiquitylation assay was performed according to the manufacturer’s instructions. Deubiquitylating activity was normalized by the control. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (B and C) MLE12 cells were transfected with Usp14-ha plasmid. (B) Forty-eight hours later, cells were treated with LPS (10 μg/ml, 3 hours). Isolated histone lysates were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (C) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. (D and E) MLE12 cells were transfected with usp14 shRNA plasmid. (D) Seventy-two hours later, cells were treated with LPS (10 μg/ml, 3 hours). Isolated histone lysates were analyzed by Western blotting with antibodies against histone H4K8ac and histone H4. (E) Determination of relative H4K8ac protein abundance by densitometric analysis of Western blots with ImageJ software. Data are means ± SEM of three independent experiments. P values were calculated by two-way ANOVA and post hoc Tukey’s test. All Western blots are representative of at least three independent experiments.
