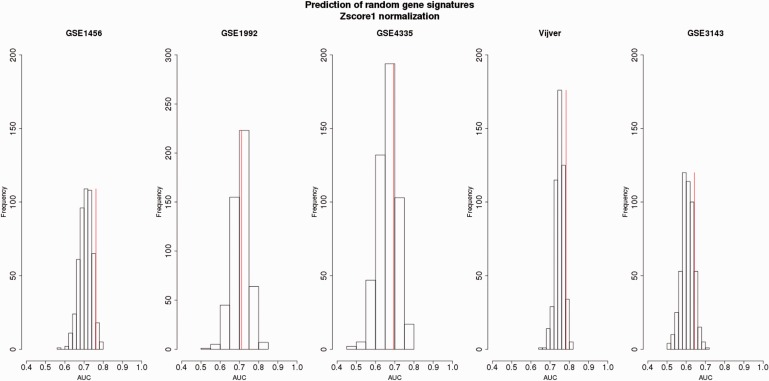
Figure 5.
Survival prediction of random gene signatures (top 100 ranked) generated from the Z -score1-normalized merged data sets with respect to the OS endpoint assessed by independent validation.Four data sets (among GSE1456, GSE1992, GSE4335, Vijver and GSE3143) were used for the training set. The data sets composing the training set were merged and adjusted by Z -score1 normalization. The top 100 random genes were derived from the training set and then validated on the remaining set (testing set) normalized by Z -score1 normalization. The testing set is indicated at the top of each plot. The prediction of non-random gene signature is illustrated by the vertical red line.For each testing data set, the average of AUCs, SD of AUCs and the percentage of AUCs higher than the AUC of the non-random gene signatures were obtained as follows:GSE1456: Mean (AUC): 0.71 ± 0.03, 6% AUCs;GSE1992: Mean (AUC): 0.70 ± 0.04, 46% AUCs;GSE4335: Mean (AUC): 0.66 ± 0.05, 25% AUCs;Vijver: Mean (AUC): 0.74 ± 0.02, 5% AUCs;GSE3143: Mean (AUC): 0.60 ± 0.03, 13% AUCs.