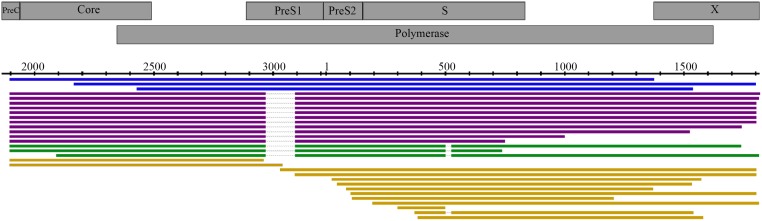
Fig 3. Alignment of nanopore reads of B6260.
Complete and partial Nanopore sequence reads were aligned with the corresponding Sanger consensus sequence used as reference. The viral core, surface, polymerase and X proteins are indicated and positions provided according to the reference sequence AM282986. Four different types of Minion reads are shown: nearly complete HBV genome (blue); reads showing a 123-nt deletion (positions 2,968–3,090) in the PreS1 region (purple); reads with the 123-nt and an additional 24-nt deletion (positions 501–524) in the “a determinant” of the S region (green); and unclassified partial reads (yellow).