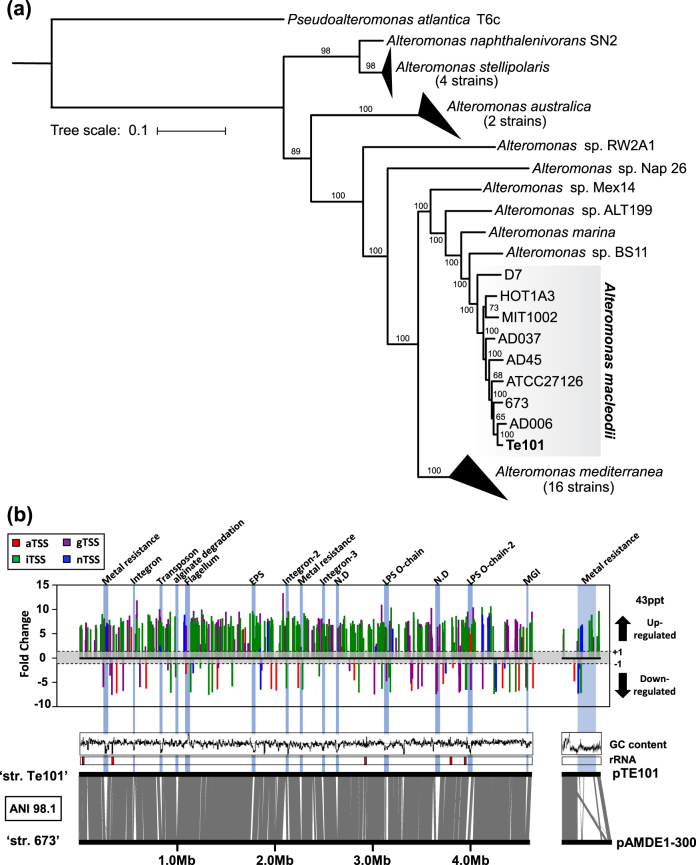
Fig. 2.
a Phylogenetic relationships between Alteromonas Te101 and other members of the genus Alteromonas. The tree is based on a concatenated set of 1015 proteins from the shared core proteome of all available Alteromonas genomes and Pseudoalteromonas atlantica T6c, which was used as an outgroup. The numbers at branches represent bootstrap values in percent. b Flexible genomic islands (fGIs) and differentially expressed TSSs. Differentially expressed TSSs are visualized on the upper panel, the y axis shows the log2-transformed fold changes. Genomic-wide synteny and fGIs (vertical blue rectangles) are shown on the lower panel. The chromosome of the A. macleodii ‘English Channel 673’ was used as a reference to detect fGIs in the Alteromonas Te101 chromosome. In the same way, the plasmid pTE101 was compared against the plasmid pAMDE1-300 of A. mediterranea strain DE1. If known, the fGI’s function is given on top. N.D represents not determined. For the list of genes within these fGIs, see Table S1