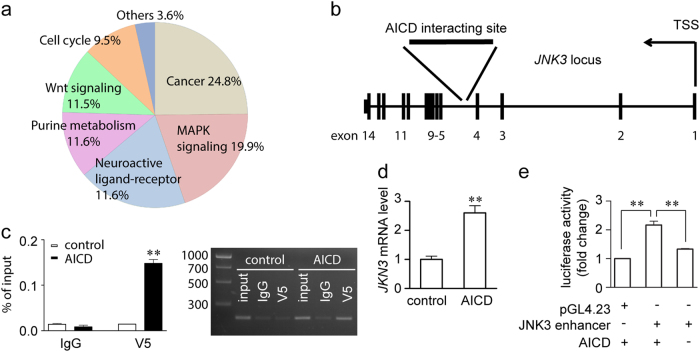
Fig. 2. Identification of JNK3 as a downstream effector of APP.
a Pathway analysis of microarray data on genes differentially expressed between control and SH-SY5Y-APP cells. b ChIP-seq screening results in SH-SY5Y cells showed that the region of AICD interaction is in the fourth intron of the JNK3 locus in chromosome 4. TSS, transcription starting site. c Validation of the ChIP-seq results. SH-SY5Y cells were transfected with either pcDNA4/V5-His or pcDNA4/V5-His-hAICD59 plasmid, and subjected to ChIP assay, using normal mouse IgG as control. Input, 5% of the sonicated chromatin. Left panel, real-time PCR results show V5, but not IgG control interacts with the JNK3 intron region (n = 3). Right panel, representative image shows the real-time PCR products resolved by agarose gel. The molecular weight of the DNA ladder shown in bp on the left. d Real-time PCR results showing the JNK3 mRNA level in pcDNA4/V5-His (control) or pcDNA4/V5-His-hAICD59 (AICD) transfected SH-SY5Y cells (n = 3). Student’s t-test: **p < 0.01. Error bars represent SEM. e Luciferase assay showing the enhancer activity of the AICD-interacting site. One-way ANOVA followed by post-hoc Bonferroni test: **p < 0.01. Error bars represent SEM