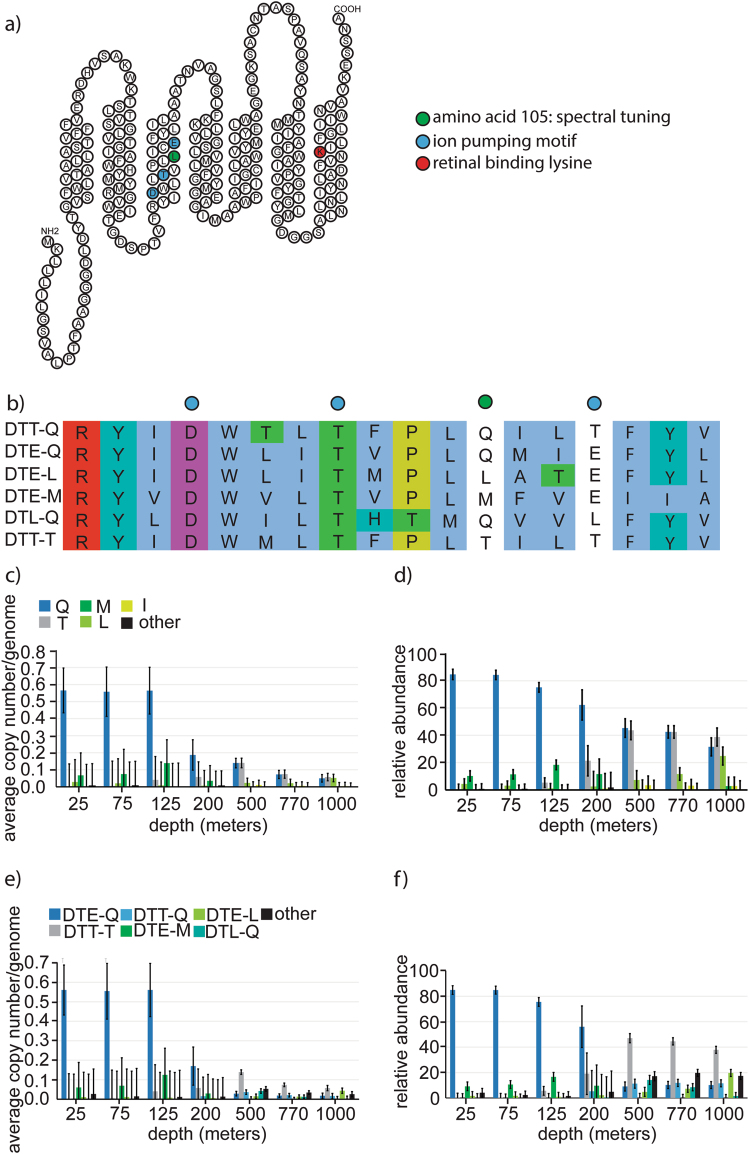
Fig. 3.
Diversity and abundance of proteorhodopsin sequence motifs. a Topology of a representative proteorhodopsin highlighting key residues in retinal binding, spectral tuning, and ion pumping. b Sequence alignment of transmembrane peptide segment three, comparing six proteorhodopsin variants highlighting variation in residues involved in spectral tuning and ion pumping. c Average copy number of rhodopsins per genome in the metagenome. d Relative abundance of spectral tuning variations in proteorhodopsins. e Average copy number per genome. f Relative abundance of variants in ion pumping motif and spectral tuning site in proteorhodopsins. c–f Blue, green, and gray color codes represent blue light absorbing, green light absorbing or unknown spectral absorption based on the spectral tuning residue of known proteorhodopsin variants. DTE is known to be a proton-pumping variant