Abstract
Molecular signatures and their interactions behind the successful establishment of infection of Mycobacterium tuberculosis (Mtb) inside macrophage are largely unknown. In this work, we present an inter-system scale atlas of the gene expression signatures, their interactions and higher order gene functions of macrophage-Mtb environment at the time of infection. We have carried out large-scale meta-analysis of previously published gene expression microarray studies andhave identified a ranked list of differentially expressed genes and their higher order functions in intracellular Mtb as well as the infected macrophage. Comparative analysis of gene expression signatures of intracellular Mtb with the in vitro dormant Mtb at different hypoxic and oxidative stress conditions led to the identification of the large number of Mtb functional groups, namely operons, regulons and pathways that were common and unique to the intracellular environment and dormancy state. Some of the functions that are specific to intracellular Mtb are cholesterol degradation and biosynthesis of immunomodulatory phenolic compounds. The molecular signatures we have identified to be involved in adaptation to different stress conditions in macrophage environment may be critical for designing therapeutic interventions against tuberculosis. And, our approach may be broadly applicable for investigating other host-pathogen interactions.
Introduction
Mycobacterium tuberculosis (Mtb) is the causative factor for tuberculosis (TB). Besides HIV, Mtb is the leading cause of death worldwide1–3. After successful infection into its host, Mtb survives as an intracellular pathogen, localized inside the macrophages and dendritic cells4,5. The phenomenon of infection of a healthy macrophage by Mtb can be divided into three inter-related stages such as triggering inflammatory response from macrophage upon infection; activation of cell mediated immunity and followed by formation of granulomas, and finally the reactivation of Mtb due to various environmental and genetic factors6. Mtb is known to survive for many years under the adverse intracellular environment of macrophages by down regulating the metabolic pathways and switching to non-replicative state7,8. It was observed that to survive in the adverse conditions, Mtb not only alters its gene expression profiles but also it alters the gene expression profiles of the host9.
Previously, many independent microarray studies were conducted to identify differentially expressed genes in Mtb as well as macrophages during the stable infection10–19. In addition to these studies, there are also few gene expression microarray studies conducted to identify adaptive mechanisms of Mtb to adverse conditions in macrophages by using in-vitro models such as nutrient starvation, hypoxia, and oxidative stress20,21. Furthermore, Meta-analysis of gene expression microarray data from independent and related datasets was also performed in several studies by which the random errors created in experiments were nullified and also the right estimate of differential expression was obtained22,23. Moreover, by performing the meta-analysis, it was also found that there is an increase in the reliability and generalization of results23–25. In this work, we performed simultaneous meta-analysis of gene expression microarray data from infected host cell (macrophage) as well as pathogen (Mtb) to identify the ranked list of gene expression signatures induced in host-pathogen environment.
Furthermore, functional over-representation analysis (FOA) of different gene groups namely complexes, pathways, regulons and gene ontology terms among the gene expression signatures was also performed which led us to identify the system-wide functions in Mtb-macrophage environment. Inclusion of operons for FOA of Mtb gene expression signatures identified many unknown pathways/complexes and adaptive mechanisms with in the macrophage environment.
Furthermore, functional over-representation analysis (FOA) of different gene groups namely complexes, pathways, regulons and gene ontology terms among the gene expression signatures was also performed which led us to identify the system-wide functions in Mtb-macrophage environment. Inclusion of operons for FOA of Mtb gene expression signatures identified many unknown pathways/complexes and adaptive mechanisms within the macrophage environment.
Materials and Methods
Preparation of gene expression microarray datasets
Gene expression microarray data sets were downloaded from two major public repository databases Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/). To remove the redundancy in the gene expression data, we have included the datasets from ArrayExpress database, if they are not found in GEO database. To facilitate the data comparison across the different platforms, gene expression microarray features were mapped to Entrez Gene IDS. Mapping information from gene names, refseq IDs, Ensembl IDs, Clone IDS, GenBank IDS to Entrez Gene IDS were obtained from various platform annotation files and also from various databases such as NCBI, BioMart, NIA array26–28.
Differential expression analysis
In order to identify differentially expressed genes during infection, we considered the samples during the final stage of the infection. Differential expression analysis of gene expression microarray data was carried out using an R package called ‘RankProd’29. The package uses a modified ‘rank product’ method29,30 to identify up or down-regulated genes in one condition against another. We have used RankProd because, it uses Rank Product method which is a non-parametric statistical method that detects genes which are consistently found among the most strongly up-regulated genes in with replicate or without replicate experiments. It is based on the assumption that under the null hypothesis the order of all items is random. Probability of finding a specific item among the top of items in a list is k/n, where k is the rank of the item and n is the total number of items. Multiplying these probabilities gives the rank product. Smaller the rank product value, smaller the probability that the observed placement of the item at the top of the lists is due to chance.
Meta-analysis of differential expression
Meta-analysis of gene expression microarrays was carried out using our previously described method31. We transformed the initial p-values of each gene obtained from the differential expression analysis of gene expression microarray datasets into the Z-scores, using the inverse cumulative distribution function (CDF)32. Then we took the weighted sum of the Z-scores and divided by the square root of sum of the weights to generate the average Z-score. Here the weights are equivalent to number of replicates in the gene expression microarray experiments. Finally the average Z-score was converted to p-value by using CDF33.
Inference of interactions of between Mtb and Human
Experimentally verified Host-Pathogen interaction data from different bacterial species were retrieved from PHISTO and HPIDB databases34,35. PHISTO is a Pathogen Host Interaction Search Tool which contains a total of 23,633 pathogen host PPI data including 9318 interactions between bacteria and human. HPIDB is a Host Pathogen Interaction Database that currently contains 23.485 interactions between 66 host and 541 pathogen species. In addition, we curated a total of 53 experimentally verified host-pathogen protein-protein interaction information from 12 pathogens including Mtb from literature. The genome sequences of Mtb and other bacteria, which have experimentally verified interactions, were downloaded from NCBI ftp site (ftp://ftp.ncbi.nlm.nih.gov/genomes/). Then, we identified orthologs of Mtb in ~1500 sequenced genomes of bacterial species, which have experimentally verified host-pathogen interactions. Finally, the experimentally verified interactions were transferred to Mtb on the basis of orthology relationship. Totally we identified 2808 host-pathogen interactions between two sets of proteins 564 from Human and 1725 from Mtb.
Preparation of functional gene groups
Annotated functions of human, i.e., pathways, motifs, gene ontology (GO) and immunological signatures were collected from Molecular Signature Database (MSigDB)36,37. Since the MSigDB does not maintain any functional gene groups for Mtb, we have curated the various functional gene groups such as complexes, pathways and operons etc., for Mtb from the published literature38.
Functional over representation analysis of gene groups
Functional over-representation analysis (FOA) was carried out using our previously described method39. For each gene in a gene group, we have converted p-value of differential expression into Z-score using, inverse normal Cumulative Distribution Function (CDF). The FOA score of each gene group is the sum of the Z-scores divided by square root of number of genes. In order to identify the overrepresented target genes of various transcription regulators (regulon), we have taken sum of the absolute value of target genes Z-scores, then the total was divided by square root of number of genes. Finally the q-value was converted to P-value by using CDF. To calibrate the gene group (containing n genes) FOA score against the background distribution, we randomly sampled the n number of Z-scores and calculated the random FOA score. We repeated this process over 100,000 times and mean and standard deviation of the sampling distribution was used for correction of original score.
Results and Discussion
Since both the host and the pathogen reciprocally influence the gene expression profiles of each other, it is pertinent to study the gene expression profiling of the pathogen as well as the host. This would provide insight into the strategies employed by the pathogen to survive in host environment and the host response to contest against the virulence of the pathogen16. Therefore, we performed the meta-analysis of gene expression changes in both Mtb and human macrophages during stable infection. In addition, we carried out Functional over-representation analysis (FOA) to identify higher order functions of induced or suppressed gene expression signatures both in Mtb as well as in the infected macrophages by considering functional groups of genes such as operons, regulons, pathways, complexes and biological process. Inclusion of diverse functional gene groups for the analysis ensured functional annotation for most of the genes. We extensively discuss the relevance of higher order functions of the genes in relation to the mechanisms adopted by the pathogen for its adaptation as well as the host defence.
Meta-analysis of differentially expressed gene signatures during infection
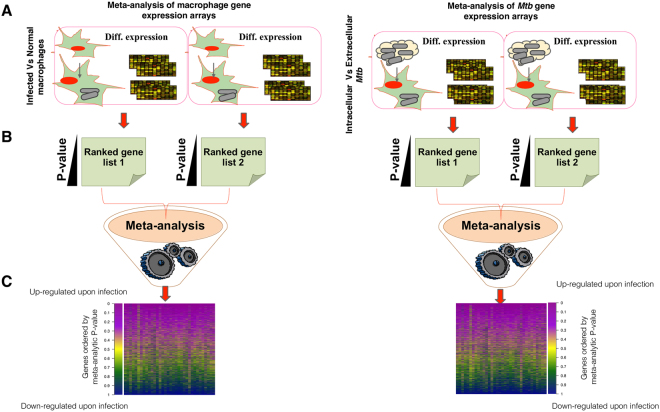
We have developed a systematic frame work to identify the genes induced in host-pathogen environment (Fig. 1). Initially, we have identified two sets of differentially expressed genes, one from the Mtb and the other from the macrophages during the course of infection. Differentially expressed genes of Mtb were identified by comparing the gene expression data of Mtb which resides inside macrophage after infection (call it intracellular Mtb) with the Mtb before infection (in vitro Mtb cultures) using RankProd29. Similarly, we have also identified differentially expressed genes of macrophages by comparing gene expression data of the infected macrophages with the uninfected macrophages (Supplementary data S1). The differential gene expression scores were given ranks and then combined to generate a single consensus ranked list, using a meta-analytical method described previously31. The genes ranked atop the consensus ranked list are more severely and consistently up-regulated during infection across experiments, and those that are ranked at the bottom were down-regulated during infection.
Figure 1.
Meta-analytic approach to identify the gene expression signatures induced in host-pathogen environment upon infection by Mtb. (A) Initially the gene expression microarray datasets of human macrophages and Mtb were collected from Gene Expression Omnibus (GEO) (B). We have used R package to generate the ranked list of differentially expressed genes by comparing the samples of infected macrophages versus normal and in vitro Mtb vs. intracellular. (C) The ranked list of differentially expressed genes were combined using previously developed meta-analytic method to generate a consensus ranked list of the differentially expressed genes in macrophages as well as in Mtb.
Host-pathogen interactions and essentiality of top ranked genes
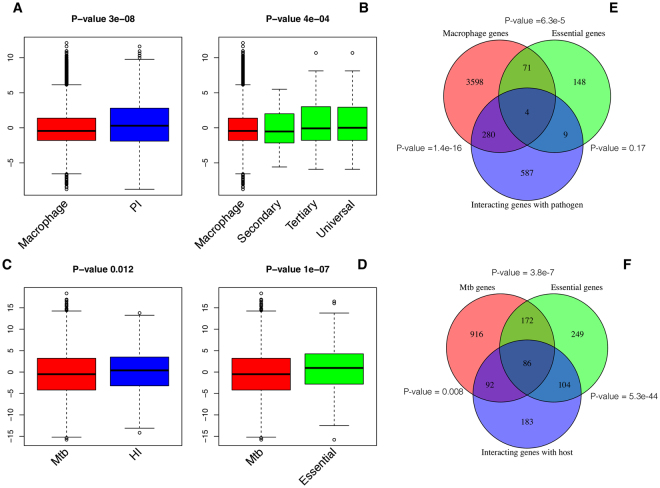
We identified orthologs of Mtb in ~1500 sequenced genomes of bacterial species, which have experimentally verified host-pathogen interactions in HPIDB and PHISTO databases as well as curated from literature34,35. The known interactions were then transferred to Mtb on the basis of orthology relationships. Totally, we identified 2808 host-pathogen interactions between two sets of proteins 564 from human and 1725 from Mtb (Supplementary data S2). Meta-analytic Z-score distribution analysis of meta-analytic ranked list from macrophage and Mtb shows that the most of the genes from both sets have above average Z-scores indicating that most of the top ranked genes encode for proteins involved in host-pathogen interactions (Fig. 2A,B). In addition, the studies conducted using genome wide siRNA screen also have large number of genes involved in the regulation of pathogen load and found to have above average Z-scores in meta-analytic ranked list of macrophage genes. (Fig. 2C)18. Moreover as the quality of RNAi hits (3 sets of genes) also have above average Z-score in the meta-analytic ranked list, indicating that top ranked genes in the meta-analytic list are highly important for defence against Mtb. Similarly, some of the genes reported to be important for survival of Mtb in different physiological environments were also have above average Z-scores (Fig. 2D)32,33,40. Overlapping statistics using hyper geometric distribution also shows that the top ranked genes from both macrophages as well as Mtb were significantly overlap with their respective set of genes that encode for proteins involved in predicted host-pathogen interactions (Fig. 2E,F). There is a significant overlap between a large number of Mtb genes that are reported to be essential and the genes, whose products are predicted to interact with the host. The common candidates between the three set Mtb genes i.e., essential, host interacting and expressed with in macrophages could be potential targets for therapeutics.
Figure 2.
Box plots showing the comparison of differential expression Z-Scores for various gene lists with overall z-scores of the genes induced in the host-pathogen environment. The positive Z-scores indicates up-regulation, whereas negative scores indicate down-regulation; (A) Z-scores of total macrophage genes that are differentially expressed upon infection and macrophage genes predicted to be pathogen interacting (PI). (B) Z-scores of total macrophage genes that are differentially expressed upon infection and macrophage genes that were reported to be essential to regulate pathogen load using different levels of siRNA screen i.e., secondary, tertiary and universal. (C) Z-scores of total Mtb genes that are differentially expressed in infected macrophages and Mtb genes predicted to be host interacting (HI). (D) Z-scores of total Mtb genes that are differentially expressed in infected macrophages and Mtb genes reported to be essential for survival in different physiological environments. P-value calculated using Wilcoxon rank sum test shows that the distributions of each pair of comparisons are significantly different. (E) Venn-diagram with P-value of overlapping statistics between the genes that are expressed with in the macrophages, pathogen interacting and essential (tertiary screen) and (F) Venn-diagram with P-value of overlapping statistics between the Mtb genes that are expressed with in the macrophages, host interacting and essential.
Higher order functions of Mtb genes induced in host-pathogen environment
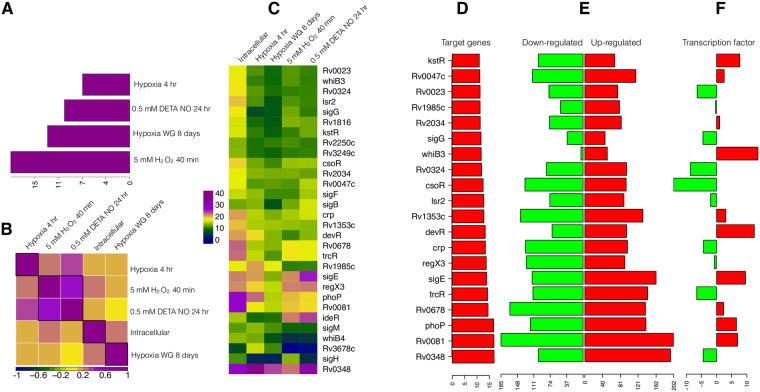
Previously there were many gene expression microarray experiments conducted to identify the adaptive mechanisms of Mtb towards the in-vitro models of nutrient starvation, hypoxia, and oxidative stress in macrophage environment. There were two types of in-vitro models studied under hypoxic conditions, one is constant oxygen (0.2%) supply model and another one is Wayne growth (WG) model, in which the oxygen supply is gradually depleted41. In order to identify the nature of environment created by macrophage towards Mtb infection, we compared the meta-analytic ranked list of gene expression signatures induced inside macrophages with a large number of previously published gene expression signatures generated using in-vitro models of diverse environmental conditions at various time points (GSE16146, GSE8786, and GSE9331)20,21,42. The gene expression signatures induced under conditions of constant O2 (0.2%) for 4hrs, Wayne growth for 8 days, 5 mM H2O2 for 40 mins and 0.5 mM DETA (Diethylenetriamine) NO for 24hrs best correlates with the meta-analytic ranked list (intracellular), indicating the likely environmental conditions Mtb exposed in macrophage environment (Fig. 3A). Gene expression signatures related to hypoxia model of WG shows stronger correlation than constant O2 (0.2%) supply, model. 5 mM H2O2 shows the strongest correlation. In addition, gene expression signatures induced in all the five different conditions show significant correlation with each other (P-value > 1.5e-08). Particularly, the conditions of hypoxia, H2O2 and NO show very strong correlation (Fig. 3B).
Figure 3.
Comparison of meta-analytic ranked list of Mtb genes induced in intracellular (macrophage) environment with the genes induced in-vitro conditions. (A) Bar plot showing the distribution of quantile normalized P-value of Spearman’s rank correlation (threshold value for significance is 2.3) between meta-analytic ranked list with genes induced under different hypoxia and oxidative stress conditions. (B) Correlation plot showing the Spearman’s rank correlation (all the correlations have significant P-value) between meta-analytic ranked list with genes induced under different hypoxia and oxidative stress conditions. (C) Heat map showing the over/under representation analysis scores for highly differentially expressed target genes of Mtb transcription regulators (score >2 is significant) under different conditions. (D) Bar plot showing the over/under representation analysis scores for highly differentially expressed target genes of various Mtb transcription regulators in intracellular conditions. (E) Divergent bar plot showing the number of down-regulated (green bars) and up-regulated genes (red bars) for each of the transcription regulator. (F) Bar plot showing the expression level of each of the transcription regulator.
In order to identify the higher order functions of Mtb genes expressed across different conditions and in intracellular environment, we carried out FOA and functional interpretation of regulons, operons and pathways as following39.
Regulons
We curated Mtb regulons by collecting known target genes of all transcription factors of Mtb from previously published literature43,44. We have performed FOA of target genes of various transcription regulators (regulons) in the ranked list of differentially expressed genes across the different conditions to get the list of the transcription regulators, which are critical for adaptation of Mtb to macrophage environment. Our analysis shows that the target genes of the persistence regulator, Rv0348 (MosR) were consistently differentially regulated across the different conditions (Fig. 3C) (Supplementary data S3). In addition, the target genes of Rv0081, PhoP, Rv0678, TrcR, SigE, RegX3, Crp, DevR, Rv1353c, Lsr2, CsoR, Rv0324, WhiB3, SigG, Rv2034, Rv1985c, Rv0023, Rv0047c, kstR were also differentially regulated in intracellular environment (Fig. 3D). Although target genes of the RegX3 and WhiB3 were significantly altered in intracellular conditions (Fig. 3E), the respective transcription regulators were not significantly differentially expressed (Fig. 3F). In contrast to the MosR expression, most of its target genes were up-regulated, indicating that it represses the majority of the genes involved in adaptation to the macrophage environment. MosR and its associated genes in an operon Rv0347-Rv0348-Rv0349 were induced during the late stage of chronic tuberculosis in mice45. PhoP is a global regulator of Mtb and it plays an important role in pathogenesis and synthesis of cell wall lipids46.
Operons
In most of the bacterial species such as Mtb, pathways, complexes and gene ontology terms were not well annotated, which hinders the functional annotation of gene expression signatures. Here, we circumvent the functional annotation problem by inclusion of predicted operons, because genes within the most of operons were known to encode proteins involved in a same pathway or potentially forming a functional protein complex.
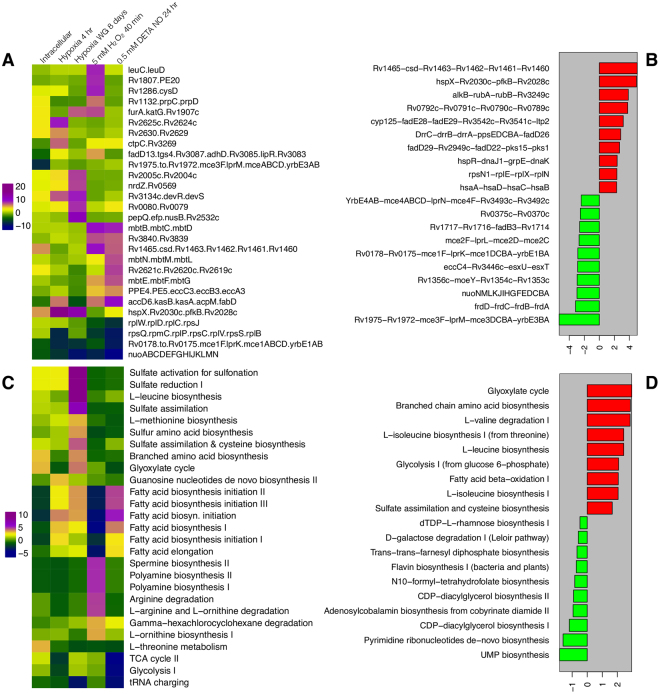
Over-representation analysis shows that the entire operon encoding for the components of NADH dehydrogenase complex-I were significantly down-regulated across the different conditions of intracellular, hypoxic and oxidative stress (Fig. 4A) (Supplementary data S4), indicating the complete shutdown of oxidative phosphorylation pathway. On the other hand, the operons such as Rv1465-csd-Rv1463-Rv1462-pps1-Rv1460, hspX-Rv2030c-pfkB-Rv2028, ceccCa1-eccB1-eccA1-espH-espG1-espF, hsaABCD, rplNXEN1, dnaK-grpE-dnaJ1-hspR, pks1-pks15-fad22-fad29, fadD26-ppsABCDE-drrABC, ltp2-fadE29-fadE28-cyp125 were highly expressed in intracellular conditions (Fig. 4B). The pps1 locus constitutes an operon with seven genes (Rv1465-Rv1460), which code for Suf-like proteins, which are part of SUF machinery, an exclusive Fe-S cluster assembly existing in Mtb47. The Pps1 protein is orthologous to SufB which is essential and conserved component of iron sulphur cluster assembly (Fe-S cluster) complex of prokaryotes. It was reported that as Fe-S clusters serve as cofactors of proteins which are involved in multiple cellular mechanisms48.
Figure 4.
Operons and pathways of Mtb with highly differentially expressed genes under different conditions. (A) Heat map showing the over/under representation scores of various operons under different stresses and intracellular (macrophage) environment. (B) Bar plot showing the operons with highly differentially expressed genes in intracellular environment. (C) Heat map showing over/under representation scores of various pathways in different conditions. (D) Bar plot showing over/under representation scores of various pathways in intracellular environments.
The operon hspX-Rv2030c-pfkB-Rv2028 was reported to be essential for the Mtb during persistent infection in the host and also for its survival during low iron, low pH and hypoxic condition49–51. The protein encoded by hspX gene of the operon is known to be highly up-regulated during the latent infection and reported to play an important role in anaerobic respiration of Mtb during stress conditions. The other genes of the operon are found to be involved in carbohydrate metabolism (pfkB) and nucleotide metabolism (Rv2030c)49–51.
The two operons hsaABCD and cyp125-fadE28-fadE29-Rv3542c-Rv3541c-ltp2 were highly conserved across the members of Mtb complex52,53. They participate in cholesterol degradation pathway and play an important role in intracellular survival of Mtb by providing cholesterol as an alternative carbon source54–59. Another two operons pks1-pks15-fad22-fad29 and fadD26-ppsABCDE-drrABC were involved in synthesis of phthiodiolone dimycocerosates (DIMs) and immuno-modulatory phenolic glycolipids, which have a role in immuno-modulation60–62. Some of the genes (drrA, drrB and drrC) in aforementioned operon were known to encode for a drug efflux pump, which confers resistance to many antibiotics63. The genes with in the operon (alkB-rubA-rubB-Rv3249c) encode for alpha-ketoglutarate-dependent dioxygenase (alkB), which catalyses oxygenation of hydrocarbons64, could be having role in cholesterol degradation. The genes involved in cholesterol utilization and other genes (Rv0792c, Rv0791c, Rv0790c, Rv0789c) with unknown function were highly expressed in intracellular environment in comparison to the other conditions.
Comparative analysis of operons with the genes induced across different conditions has identified several functions that are common and unique to each of the environment. Pathways involved in biosynthesis of iron-sulfur cluster, mycobactins (mbtB-mbtC-mbtD) and meromycolic acids (accD6-kasB-kasA-acpB-fabD) were highly expressed in presence of NO and H2O2 indicating their role in oxidative stress defence. On the other hand the genes with in the operons (devR-devS-Rv3134c and hspX-Rv2030c-pfkB-Rv2028c) were highly expressed during hypoxic conditions. Furthermore the genes with in the operons (furA-katG-Rv1907c and ahpC-ahpD) were expressed across intracellular, hypoxic, H2O2 induced oxidative stress conditions. In contrast to the previous reports, co-expression of katG and furA indicates that furA may not be a repressor of katG under these conditions65.
Pathways
Over-representation analysis of various Mtb pathways among the genes induced during infection shows that the glyoxylate pathway which is known to play an important role in persistence of Mtb in macrophages66 is only active in Wane growth model of hypoxia and intracellular conditions (Fig. 4C) (Supplementary data S4). In addition, pathways involved in the biosynthesis of chorismate and branched chain amino acids such as L-leucine, L-isoleucine (from threonine) and assimilation of sulfate and cysteine biosynthesis were highly expressed during hypoxia and intracellular conditions, but not during oxidative stress. Furthermore, various catabolic pathways such as fatty acid & beta-oxidation I, glycolysis I (from glucose 6-phosphate), and glyoxylate cycles are highly expressed during intracellular conditions (Fig. 4D). The chorismate pathway, also called as shikimate pathway is found in Mtb but absent in humans and other mammals67,68. This pathway produces chorismate, which is a key intermediate compound for the biosynthesis of amino acids, ubiquinones, siderophores and other important metabolites essential for Mtb growth and survival in the host69–73. Overall, the pathways involved in the biosynthesis of L-Leucine, chorismate, cysteine, and assimilation of sulfate are linked to the long-time survival of Mtb during the pathogenesis.
Higher order functions of differentially expressed macrophages genes upon Mtb infection
Pathways
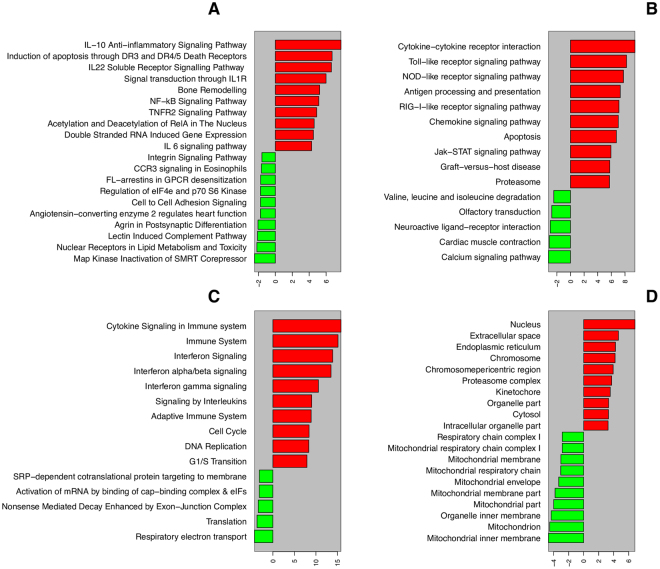
In order to identify the macrophage pathways that were induced during infection, we have carried out over-representation analysis of pathway genes from various databases such as BioCarta, KEGG, and Reactome74–76 (Fig. 5) (Supplementary data S5). The BioCarta pathways namely, IL-10 anti-inflammatory signalling, induction of apoptosis through DR3 and DR4/5 Death Receptors, IL22 soluble receptor signalling, NF-kB, TNFR2 signalling and Acetylation/deacetlyation of RelA were found to be highly expressed (Fig. 5A).
Figure 5.
Macrophage pathways and cellular components with highly differentially expressed genes upon infection. (A,B and C) shows over/under represented pathways from Biocarta, KEGG and Reactome, respectively. (D) shows the over/under represented cellular components.
IL-10 family of cytokines has a total of 6 IL’s. In contrast to the rest of the IL-10 family of cytokines, IL-10 acts as an anti-inflammatory cytokine and is also capable of inhibiting the synthesis of pro-inflammatory cytokines such as IL-2, TNF-α, IL-3, IFN-α which indirectly leads to the survival of Mtb inside the infected macrophage. Supporting this fact, there are reported evidence from the infection studies of Mycobacterium avium sub species paratuberclosis (MAP). It was documented that MAP adopts a different strategy to escape from the host defence. MAP activates the mitogen activated protein kinase pathway, which finally results in up-regulation of IL-10 and repression of other inflammatory cytokines. Further the up-regulated IL-10 also inhibits the apoptosis in macrophages6,77,78. According to our findings and the supporting evidence, it may be assumed that the up-regulation of IL-10, an anti-inflammatory pathway of macrophage may aid in the growth and virulence of Mtb and may also be considered as one of the immune-modulatory strategies adopted my Mtb to escape from host defence mechanisms for its survival.
The pathway ‘induction of apoptosis through the engagement of death receptors’ has a role in apoptosis of infected macrophages via engagement of variety of death receptors such as Fas, TNFR, DR3, DR4 and DR579. Apoptosis of this kind, where there is engagement of death receptors is the sign of innate immune response of macrophages against Mtb79. IL22 soluble receptor signalling pathway was reported to inhibit the growth of Mtb by enhancing the expression of intracellular signalling molecule, calgranulin A. In addition, it was reported to be involved in tissue modulation, induction of proliferative and anti-apoptotic pathways and production of anti-microbial substances80.
NF-kB pathway, inflammatory cytokine and TNF-α are responsible for inflammatory response towards Mtb infection81. TNF- α shows its immune response in variety of ways such as macrophage activation, induction of cytokines, chemokines and induction of apoptosis and also restriction of bacterial growth in infected macrophage81–85. TNFR2 signalling pathway is mainly responsible for apoptosis, acetylation/deacetlyation of RelA and is responsible for innate immune response against Mtb. Overall the over-representation analysis in Biocarta pathway shows that all the pathways related to recognition of molecular patterns of Mtb are responsible for triggering inflammation and apoptosis, are up-regulated.
Over-representation of analysis of KEGG pathways show that the genes involved in cytokine-cytokine receptor interaction, toll-like receptor signalling, NOD-like receptor signalling, RIG-I like signalling, chemokine signalling and apoptosis signalling were highly up-regulated in the macrophages during Mtb infection (Fig. 5B). After infection of Mtb in pulmonary alveoli, macrophages express pattern recognition receptors (PRR) which in turn recognize the pathogen associated molecular patterns (PAMPS). The recognition of PAMPS by PRRs leads to the activation of transcription factors, which are involved in providing innate immunity86. In the case of mycobacterial infections, mycobacterial cell wall-associated components such as lipoarabinomannan are recognized by TLR1 and TLR2. Similarly there are several cell wall associated components which may be recognized during the time of mycobacterial infection as reported earlier87–89. In NOD-like receptor signalling pathway, the NOD like receptors belonging to NLR family and they may play key roles in innate immunity and inflammation90. They recognize the peptidoglycan fragments of the pathogens escaped from endosomal compartments leading to the activation of cytokine production and ultimately resulting in programmed cell death91,92.
Apart from the NOD like receptors, there are also few other proteins called as cytosolic RNA helicases (RIG-I like signalling pathway) that were also highly up-regulated. These cytosolic RNA helicases (RIG-I-like receptors) are reported to recognize the antigens from viral pathogens and induce the innate immune response against them by triggering the signalling pathways which produce the type I interferon and inflammatory cytokines93,94. These have been reported to be involved in the intracellular detection of all pathogens including Mtb95. Chemokine signalling pathway plays an important role in migration of leukocytes to the site of infection and orchestrates the host defence mechanism96.
The genes involved in apoptotic pathways were highly up-regulated (Fig. 5A,B). The cells follow two different kinds of pathways for undergoing apoptosis: the death receptor or extrinsic pathway and the mitochondrial or intrinsic pathway, where the final dead cells are cleared through phagocytosis by the macrophages97–99. In addition, the pathways in involved in branched chain amino-acid degradation were down-regulated, unlike in intracellular Mtb, in which the biosynthetic pathways of branched chain amino-acids were up-regulated as mentioned previously.
Analysis of Reactome pathways show that there are two kinds of the pathways that were up-regulated during infection: pathways pertaining to immune system activation and pathways responsible for cell survival and growth. The former category consists of cytokine signalling related genes, interferon signalling, interleukins signalling and adaptive immunity. The second category consisting of the pathways related to cell cycle, DNA replication and G1/S transition pathways were also observed to be highly up-regulated (Fig. 5C). In the first category, most of the pathways which were highly regulated were found to be cytokines with diverse functions. As previously described, cytokines have immune response triggering properties against the infections in the host. Interferon’s are also cytokines (pleiotropic cytokines) which have immune-regulatory properties 103. In our analysis, we also have found the pathways pertaining to interferon α/β/γ signalling as highly up-regulated which are also actively involved in triggering immune response. These results confirm that the pathways in the first category are involved in providing innate and adaptive immunity against infection. The pathways which are involved in cell cycle, DNA replication, G1/S transition are well documented to be linked with cell growth and survival.
Cellular components
Over-representation analysis of genes encoding various cellular components shows that the genes that encode for proteins localized to Endoplasmic Reticulum (ER) were observed to be up-regulated during infection. In macrophages, the mycobacterial antigen ESAT-6 evokes the ER stress, which in turn mediates the process of apoptosis100. This induction of apoptosis further leads to the killing of Mtb. The other highly overrepresented proteins in the cellular localization data set are mainly related to the intracellular proteins associated with kinetochore (Fig. 5D) (Supplementary data S5). In contrast to the ER, most of the mitochondrial components were down-regulated, indicating the shutdown of most of the mitochondrial functions in infected macrophages. Mtb virulence is known to correlate with mitochondrial cytochrome c release in infected macrophages101. Mitochondria could be an important target for mycobacteria, because of its involvement in the regulation of immune response and cell survival/programmed cell death102–104. Furthermore, the Mtb could also be exploiting some of the mitochondrial functions considering that mitochondria derived from endosymbiosis of an ancestral bacteria105. It has been shown that mycobacterial proteins carry mitochondrial localization signals, which may help its virulence proteins to be transported to the mitochondria and further hijack it’s functions106.
Conclusions
In this report by performing the meta-analysis of publicly available gene expression datasets and analysis of higher order functions and interactions, we have derived comprehensive view about cross talk between Mtb and macrophages during the stable infection. We observed a strong correlation of gene expression signatures between the intracellular bacteria with in-vitro models of dormancy conditions, which reflects the likely environmental conditions in existing in macrophages at the time of infection. Moreover from this analysis, it is clear that Mtb effectively fights against the oxidative, starvation and hypoxic conditions presented by the host as defence against infection. Analysis of different functional signatures associated with different stress conditions provides the valuable information related to the strategies adapted by Mtb inside the macrophage for its survival and persistence. For instance such as, the mechanisms adapted by Mtb to encounter the oxidative stress induced by intracellular H2O2, through catalase-peroxidase and alkyl hydro peroxide reductase, similarly, recruitment of iron-sulfur clusters, mycobactins and meromycolic acids to encounter the oxidative stress induced by NO shows the strategies adapted by Mtb to survive in infected macrophage. Further, from our analysis it is observed that, Mtb effectively tackles the harsh environmental conditions of macrophage by intracellular environment by up-regulation of cholesterol utilization pathways and bio-synthesis of immuno-modulatory phenolic glycolipids. Further study of these pathways and other genes with unknown function would establish their role and mechanisms of action to counter each of the stress conditions posed by macrophage environment. In addition possible pathways of Mtb that target and exploit mitochondria should be identified. This study clearly inferred that, when therapeutic strategies to combat dormant Mtb should be designed, the multiple pathways involved in adaptation to different stress conditions and pathways that target mitochondrial functions should be taken into consideration.
Electronic supplementary material
Acknowledgements
This work was supported by Defence Research & Development Establishment [DRDE/TC/05414/Proj/Task-211/14 to S.Y.]; Department of Science Technology-Science and Engineering Research Board (DST-SERB) [SB/YS/LS‐230/2013 to S.Y.]; National Institute of Animal Biotechnology to S.Y.; DST-SERB [SB/YS/LS-125/2014 to Y.A.] and DST-SERB Post-doctoral Fellowship [PDF/2015/000313 to PVPSA].
Author Contributions
S.Y. designed the study; P.V.P.S.A., S.K.M., A.R. and S.Y. collected the data; S.Y. carried out the computation and generated Figures; P.V.P.S.A., Y.A., S.K.M., S.K. and S.Y. analysed the data; P.V.P.S.A., Y.A. and S.Y. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
P. V. Parvati Sai Arun and Sravan Kumar Miryala contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22884-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Daniel TM. The history of tuberculosis. Respiratory medicine. 2006;100:1862–1870. doi: 10.1016/j.rmed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Gideon HP, Flynn JL. Latent tuberculosis: what the host “sees. Immunologic research. 2011;50:202–212. doi: 10.1007/s12026-011-8229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Organization, W. H. Global tuberculosis report 2015. (World Health Organization, 2015).
- 4.Bloom, B. R. Tuberculosis: pathogenesis, protection, and control. (ASM press, 1994).
- 5.Snider, D., Raviglione, M., Kochi, A. & Bloom, B. Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, DC (1994).
- 6.Sasindran SJ, Torrelles JB. Mycobacterium Tuberculosis Infection and Inflammation: what is Beneficial for the Host and for the Bacterium? Frontiers in microbiology. 2011;2:2. doi: 10.3389/fmicb.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta UD, Katoch VM. Understanding the phenomenon of persistence in mycobacterial infections. Indian J Lepr. 1997;69:385–393. [PubMed] [Google Scholar]
- 9.Rana A, Ahmed M, Rub A, Akhter Y. A tug-of-war between the host and the pathogen generates strategic hotspots for the development of novel therapeutic interventions against infectious diseases. Virulence. 2015;6:566–580. doi: 10.1080/21505594.2015.1062211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubnau E, Smith I. Mycobacterium tuberculosis gene expression in macrophages. Microbes and infection. 2003;5:629–637. doi: 10.1016/S1286-4579(03)00090-X. [DOI] [PubMed] [Google Scholar]
- 11.Fu YR, Gao KS, Ji R, Yi ZJ. Differential transcriptional response in macrophages infected with cell wall deficient versus normal Mycobacterium Tuberculosis. International journal of biological sciences. 2015;11:22–30. doi: 10.7150/ijbs.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendall SL, Rison SC, Movahedzadeh F, Frita R, Stoker NG. What do microarrays really tell us about M. tuberculosis? Trends in microbiology. 2004;12:537–544. doi: 10.1016/j.tim.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Ragno S, et al. Changes in gene expression in macrophages infected with Mycobacterium tuberculosis: a combined transcriptomic and proteomic approach. Immunology. 2001;104:99–108. doi: 10.1046/j.1365-2567.2001.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnappinger D, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. The Journal of experimental medicine. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tailleux L, et al. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PloS one. 2008;3:e1403. doi: 10.1371/journal.pone.0001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volpe E, et al. Gene expression profiling of human macrophages at late time of infection with Mycobacterium tuberculosis. Immunology. 2006;118:449–460. doi: 10.1111/j.1365-2567.2006.02378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS pathogens. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar D, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Thuong NT, et al. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS pathogens. 2008;4:e1000229. doi: 10.1371/journal.ppat.1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PloS one. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Normand SL. Meta-analysis: formulating, evaluating, combining, and reporting. Statistics in medicine. 1999;18:321–359. doi: 10.1002/(SICI)1097-0258(19990215)18:3<321::AID-SIM28>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 23.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS medicine. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson L. External validity, generalizability, and knowledge utilization. Journal of nursing scholarship: an official publication of Sigma Theta Tau International Honor Society of Nursing. 2004;36:16–22. doi: 10.1111/j.1547-5069.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer research. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 26.Guberman JM, et al. BioMart Central Portal: an open database network for the biological community. Database: the journal of biological databases and curation. 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haider S, et al. BioMart Central Portal–unified access to biological data. Nucleic acids research. 2009;37:W23–27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 29.Hong F, et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 30.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 31.Cinghu S, et al. Integrative framework for identification of key cell identity genes uncovers determinants of ES cell identity and homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1581–1590. doi: 10.1073/pnas.1318598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Molecular microbiology. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 33.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS pathogens. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar R, Nanduri B. HPIDB-a unified resource for host-pathogen interactions. BMC bioinformatics. 2010;11:S16. doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tekir SD, et al. PHISTO: pathogen–host interaction search tool. Bioinformatics. 2013;29:1357–1358. doi: 10.1093/bioinformatics/btt137. [DOI] [PubMed] [Google Scholar]
- 36.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic acids research. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arun P, et al. Identification and functional analysis of essential, conserved, housekeeping and duplicated genes. FEBS letters. 2016;590:1428–1437. doi: 10.1002/1873-3468.12192. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YJ, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS pathogens. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infection and immunity. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Frontiers in microbiology. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanz J, et al. The transcriptional regulatory network of Mycobacterium tuberculosis. PloS one. 2011;6:e22178. doi: 10.1371/journal.pone.0022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turkarslan S, et al. Network portal: a database for storage, analysis and visualization of biological networks. Nucleic acids research. 2014;42:D184–190. doi: 10.1093/nar/gkt1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talaat AM, et al. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. Journal of bacteriology. 2007;189:4265–4274. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walters SB, et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Molecular microbiology. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 47.Huet G, Daffe M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen’s survival. Journal of bacteriology. 2005;187:6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roche B, et al. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochimica et biophysica acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Mushtaq K, et al. Rv2031c of Mycobacterium tuberculosis: a master regulator of Rv2028-Rv2031 (HspX) operon. Frontiers in microbiology. 2015;6:351. doi: 10.3389/fmicb.2015.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman DR, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan Y, Crane DD, Barry CE., 3rd Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. Journal of bacteriology. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capyk JK, et al. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. The Journal of biological chemistry. 2009;284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Fernandez E, et al. A highly conserved mycobacterial cholesterol catabolic pathway. Environ Microbiol. 2013;15:2342–2359. doi: 10.1111/1462-2920.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang JC, Harik NS, Liao RP, Sherman DR. Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. The Journal of infectious diseases. 2007;196:788–795. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- 55.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang M, Guja KE, Thomas ST, Garcia-Diaz M, Sampson NS. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS chemical biology. 2014;9:2632–2645. doi: 10.1021/cb500232h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dresen C, et al. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. The Journal of biological chemistry. 2010;285:22264–22275. doi: 10.1074/jbc.M109.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouellet H, Johnston JB, de Montellano PR. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends in microbiology. 2011;19:530–539. doi: 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Geize R, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Constant P, et al. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. The Journal of biological chemistry. 2002;277:38148–38158. doi: 10.1074/jbc.M206538200. [DOI] [PubMed] [Google Scholar]
- 61.Pang JM, et al. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. Journal of bacteriology. 2012;194:715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vergnolle O, et al. Biosynthesis of cell envelope-associated phenolic glycolipids in Mycobacterium marinum. Journal of bacteriology. 2015;197:1040–1050. doi: 10.1128/JB.02546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choudhuri BS, et al. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. The Biochemical journal. 2002;367:279–285. doi: 10.1042/bj20020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Churchill SA, Harper JP, Churchill PF. Isolation and characterization of a Mycobacterium species capable of degrading three- and four-ring aromatic and aliphatic hydrocarbons. Appl Environ Microbiol. 1999;65:549–552. doi: 10.1128/aem.65.2.549-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zahrt TC, Song J, Siple J, Deretic V. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Molecular microbiology. 2001;39:1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 66.McKinney JD, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 67.Bentley R. The shikimate pathway–a metabolic tree with many branches. Critical reviews in biochemistry and molecular biology. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 68.Mdluli K, Spigelman M. Novel targets for tuberculosis drug discovery. Current opinion in pharmacology. 2006;6:459–467. doi: 10.1016/j.coph.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 69.Prakash P, Pathak N, Hasnain SE. pheA (Rv3838c) of Mycobacterium tuberculosis encodes an allosterically regulated monofunctional prephenate dehydratase that requires both catalytic and regulatory domains for optimum activity. The Journal of biological chemistry. 2005;280:20666–20671. doi: 10.1074/jbc.M502107200. [DOI] [PubMed] [Google Scholar]
- 70.Yellaboina S, Ranjan S, Vindal V, Ranjan A. Comparative analysis of iron regulated genes in mycobacteria. FEBS letters. 2006;580:2567–2576. doi: 10.1016/j.febslet.2006.03.090. [DOI] [PubMed] [Google Scholar]
- 71.Dosselaere F, Vanderleyden J. A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Critical reviews in microbiology. 2001;27:75–131. doi: 10.1080/20014091096710. [DOI] [PubMed] [Google Scholar]
- 72.Parish T, Stoker NG. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology. 2002;148:3069–3077. doi: 10.1099/00221287-148-10-3069. [DOI] [PubMed] [Google Scholar]
- 73.Schneider CZ, Parish T, Basso LA, Santos DS. The two chorismate mutases from both Mycobacterium tuberculosis and Mycobacterium smegmatis: biochemical analysis and limited regulation of promoter activity by aromatic amino acids. Journal of bacteriology. 2008;190:122–134. doi: 10.1128/JB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Croft, D. et al. Reactome: a database of reactions, pathways and biological processes. Nucleic acids research, gkq1018 (2010). [DOI] [PMC free article] [PubMed]
- 75.Kanehisa M. The KEGG database. silico simulation of biological processes. 2002;247:91–103. doi: 10.1002/0470857897.ch8. [DOI] [Google Scholar]
- 76.Nishimura D. BioCarta. Biotech Software & Internet Report: The Computer Software Journal for Scient. 2001;2:117–120. doi: 10.1089/152791601750294344. [DOI] [Google Scholar]
- 77.Hussain T, Shah SZ, Zhao D, Sreevatsan S, Zhou X. The role of IL-10 in Mycobacterium avium subsp. paratuberculosis infection. Cell communication and signaling: CCS. 2016;14:29. doi: 10.1186/s12964-016-0152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoo KH, Kim SK, Chung JH, Chang SG. Association of IL10, IL10RA, and IL10RB polymorphisms with benign prostate hyperplasia in Korean population. J Korean Med Sci. 2011;26:659–664. doi: 10.3346/jkms.2011.26.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li J, Chai QY, Liu CH. The ubiquitin system: a critical regulator of innate immunity and pathogen-host interactions. Cellular & molecular immunology. 2016;13:560–576. doi: 10.1038/cmi.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karpov LM. [The role of Na K ATPase in thiamine and lipoic acid interrelations during their absorption in the gastrointestinal tract of mice] Fiziologicheskii zhurnal. 1989;35:51–57. [PubMed] [Google Scholar]
- 81.Fallahi-Sichani M, Kirschner DE, Linderman JJ. NF-kappaB Signaling Dynamics Play a Key Role in Infection Control in Tuberculosis. Frontiers in physiology. 2012;3:170. doi: 10.3389/fphys.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Algood HM, et al. TNF influences chemokine expression of macrophages in vitro and that of CD11b+ cells in vivo during Mycobacterium tuberculosis infection. Journal of immunology. 2004;172:6846–6857. doi: 10.4049/jimmunol.172.11.6846. [DOI] [PubMed] [Google Scholar]
- 83.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 84.Gutierrez MG, et al. NF-kappa B activation controls phagolysosome fusion-mediated killing of mycobacteria by macrophages. Journal of immunology. 2008;181:2651–2663. doi: 10.4049/jimmunol.181.4.2651. [DOI] [PubMed] [Google Scholar]
- 85.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews. Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanchez D, et al. Role of TLR2- and TLR4-mediated signaling in Mycobacterium tuberculosis-induced macrophage death. Cell Immunol. 2010;260:128–136. doi: 10.1016/j.cellimm.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 87.Bulut Y, et al. Mycobacterium tuberculosis heat shock proteins use diverse Toll-like receptor pathways to activate pro-inflammatory signals. The Journal of biological chemistry. 2005;280:20961–20967. doi: 10.1074/jbc.M411379200. [DOI] [PubMed] [Google Scholar]
- 88.Means TK, et al. Human toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. Journal of immunology. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 89.Tapping RI, Tobias PS. Mycobacterial lipoarabinomannan mediates physical interactions between TLR1 and TLR2 to induce signaling. J Endotoxin Res. 2003;9:264–268. doi: 10.1177/09680519030090040801. [DOI] [PubMed] [Google Scholar]
- 90.Benko S, Philpott DJ, Girardin SE. The microbial and danger signals that activate Nod-like receptors. Cytokine. 2008;43:368–373. doi: 10.1016/j.cyto.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 91.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunological reviews. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang XL, et al. Bivariate whole genome linkage analyses for total body lean mass and BMD. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2008;23:447–452. doi: 10.1359/jbmr.071033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Current opinion in immunology. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 94.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 95.Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wong MM, Fish EN. Chemokines: attractive mediators of the immune response. Seminars in immunology. 2003;15:5–14. doi: 10.1016/S1044-5323(02)00123-9. [DOI] [PubMed] [Google Scholar]
- 97.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer biology & therapy. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 99.Savitskaya MA, Onishchenko GE. Mechanisms of Apoptosis. Biochemistry. Biokhimiia. 2015;80:1393–1405. doi: 10.1134/S0006297915110012. [DOI] [PubMed] [Google Scholar]
- 100.Choi HH, et al. Endoplasmic reticulum stress response is involved in Mycobacterium tuberculosis protein ESAT-6-mediated apoptosis. FEBS letters. 2010;584:2445–2454. doi: 10.1016/j.febslet.2010.04.050. [DOI] [PubMed] [Google Scholar]
- 101.Abarca-Rojano E, Rosas-Medina P, Zamudio-Cortez P, Mondragon-Flores R, Sanchez-Garcia FJ. Mycobacterium tuberculosis virulence correlates with mitochondrial cytochrome c release in infected macrophages. Scand J Immunol. 2003;58:419–427. doi: 10.1046/j.1365-3083.2003.01318.x. [DOI] [PubMed] [Google Scholar]
- 102.Lobet E, Letesson JJ, Arnould T. Mitochondria: a target for bacteria. Biochem Pharmacol. 2015;94:173–185. doi: 10.1016/j.bcp.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 103.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahajan G, Mande SC. Using structural knowledge in the protein data bank to inform the search for potential host-microbe protein interactions in sequence space: application to Mycobacterium tuberculosis. BMC Bioinformatics. 2017;18:201. doi: 10.1186/s12859-017-1550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nass S, Nass MM. Intramitochondrial Fibers with DNA Characteristics. Ii. Enzymatic and Other Hydrolytic Treatments. J Cell Biol. 1963;19:613–629. doi: 10.1083/jcb.19.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rana A, Kumar D, Rub A, Akhter Y. Proteome-scale identification and characterization of mitochondria targeting proteins of Mycobacterium avium subspecies paratuberculosis: Potential virulence factors modulating host mitochondrial function. Mitochondrion. 2015;23:42–54. doi: 10.1016/j.mito.2015.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.