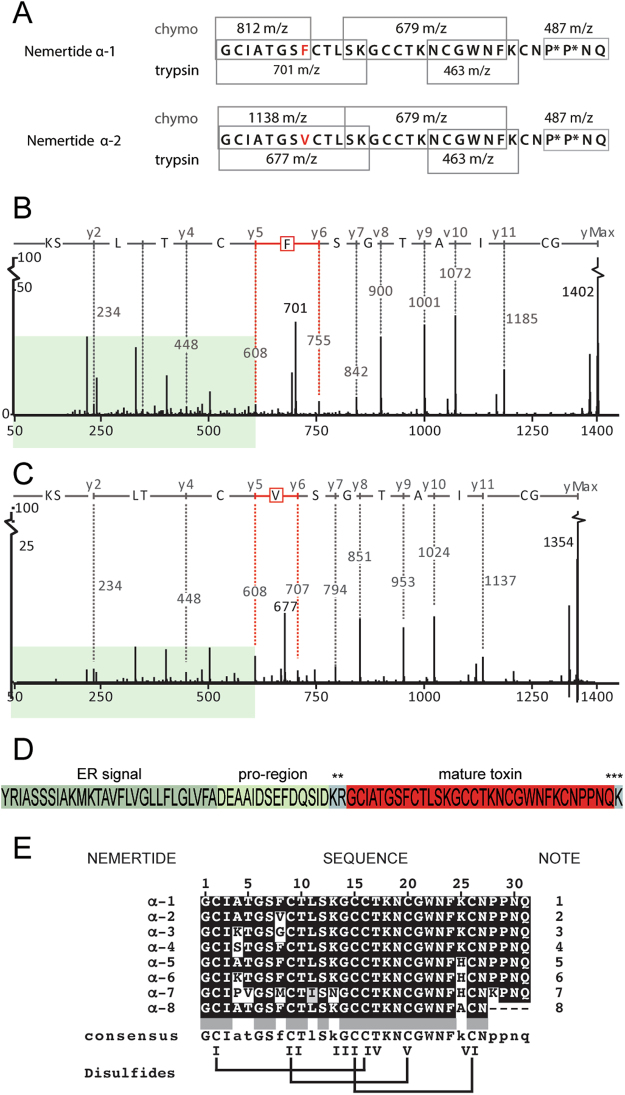
Figure 3.
Sequencing and transcriptomic identification of α-nemertides. (A) Ion-map of enzymatically cleaved peptides sequenced by MSMS, *hydroxyproline. The C-terminal tryptic fragment (residues 26–31) of α-1 and α-2 did not give any sequence by MSMS. The 487 m/z fragment is consistent with P*P*NQ, which was found in both the native α-1 and -2 and confirmed by synthetic α-1. (B) and (C) Comparison of the N-terminal tryptic fragments of nemertides α-1 and α-2 demonstrate the change of F into V at position 8; the b and y ion series where peaks overlap are highlighted in pale green. Position 8 is highlighted in red in both spectra. All Cys residues were alkylated using iodoacetamide (Cys + 57). (D) α-1-precursor with the ER-signal, pro-region and mature toxin marked (predicted by Conoprec38. **Pre-sequence cleavage site, ***post-sequence lysine cleavage. (E) α-nemertides found in available transcriptomes. Disulfide connectivities in the sequences are inferred from the nemertide α-1. Notes: 1 present in Lineus longissimus, L. lacteus and L. ruber; 2 L. longissimus, L. ruber; 3 L. lacteus, L. pseudolacteus; 4 L. sanguieneus; 5 L. pseudolacteus; 6 L. sanguieneus; 7 L. ruber. One partial sequence was found in R. occultus, 8.