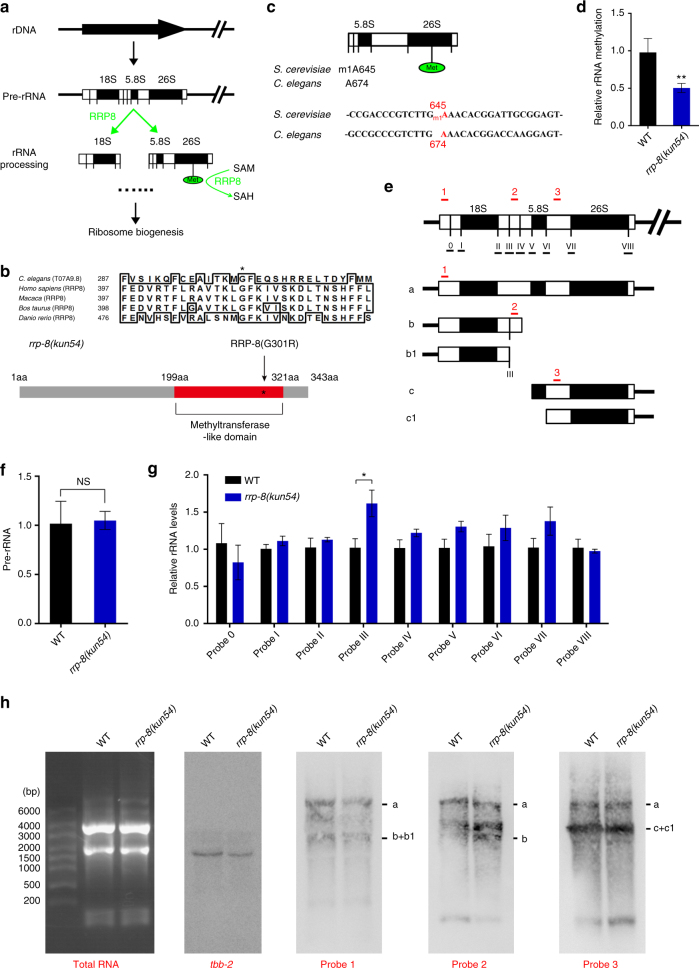
Fig. 2.
Mutation of RRP-8 impairs rRNA methylation and processing. a Schematic diagram of ribosome biogenesis in yeast and mammals. RRP8 may be involved in pre-rRNA cleavage and methylation. b Top panel: alignment of partial amino acid sequences of RRP-8 across several organisms, including Homo sapiens (NP_056139.1), Macaca mulatta (AFH30501.1), Bos taurus (NP_001179303.1), and Danio rerio (XP_017206635); the conserved glycine is marked by an asterisk. Bottom panel: schematic diagram of the mutation (G301R) in the methyltransferase-like domain (MLD) of RRP-8 and the mutation site is indicated by an asterisk. c The m1A modification site of 25/26S rRNA in yeast and C. elegans. d The rRNA methylation rate at the indicated site (A674) in 26S rRNA. e Schematic diagram of the pre-rRNA (a) and its processed intermediates (b, b1, c, and c1) in C. elegans14, 22. The cleavage sites (0–VIII) are indicated in the pre-rRNA. The underlined “_” under 0–VIII represents QPCR regions for detection of the splicing efficiency. The positions of probes 1–3 used for northern blot analysis are shown in the indicated regions (red “_”). f, g Detection of Pre-rRNA levels (f) and splicing efficiency of probable cleavage sites in pre-rRNA (g) by RT-qPCR. The levels of rRNA at each indicated site (0–VIII) were detected in 1-day-old WT and rrp-8(kun54) worms by RT-qPCR with the corresponding primers listed in the corresponding method. Data are presented as the mean 2−ddCt ± SD of four biological replicates using tbb-2 as a reference gene. h Northern blot analysis of pre-rRNA and its processed intermediates in WT and rrp-8(kun54) worms. Equal amounts of total RNA (5 μg) were loaded on a 1.2% denaturing formaldehyde/agarose gel and analyzed by northern blotting with digoxin-labeled probes 1–3 indicated in e. tbb-2 was used as a control of RNA quality. Data are presented as the mean ± SD of four biological repeats. Significant difference between WT and the rrp-8(kun54) mutant, Student’s t-test, **P < 0.01, *P < 0.05. NS, no significant difference