Abstract
Recent study has shown that RNA interference (RNAi) is efficient in emerald ash borer (EAB), Agrilus planipennis, and that ingestion of double-stranded RNA (dsRNA) targeting specific genes causes gene silencing and mortality in neonates. Here, we report on the identification of highly effective target genes for RNAi-mediated control of EAB. We screened 13 candidate genes in neonate larvae and selected the most effective target genes for further investigation, including their effect on EAB adults and on a non-target organism, Tribolium castaneum. The two most efficient target genes selected, hsp (heat shock 70-kDa protein cognate 3) and shi (shibire), caused up to 90% mortality of larvae and adults. In EAB eggs, larvae, and adults, the hsp is expressed at higher levels when compared to that of shi. Ingestion of dsHSP and dsSHI caused mortality in both neonate larvae and adults. Administration of a mixture of both dsRNAs worked better than either dsRNA by itself. In contrast, injection of EAB.dsHSP and EAB.dsSHI did not cause mortality in T. castaneum. Thus, the two genes identified cause high mortality in the EAB with no apparent phenotype effects in a non-target organism, the red flour beetle, and could be used in RNAi-mediated control of this invasive pest.
Introduction
RNA interference (RNAi) is a specific gene-silencing mechanism that is being developed as a tool in pest management programs to protect plants against insect pests. Because its mode of action involves silencing specific target genes based on complementary sequences, RNAi is considered more specific than broad-spectrum and conventional pesticides, decreasing the chances of collateral damage and negative effects on non-target and beneficial organisms1. In addition, the diversity of Dicer-substrate siRNAs produced after dsRNA cleavage may impede the evolution of RNAi resistance due to polymorphism in the nucleotide sequences, thereby increasing the durability of RNAi technology in the field2. Utilization of RNAi technology to protect agricultural crops is more advanced than in horticultural crops or forestry3–6.
The efficacy of RNAi varies among insects, and several critical factors may be responsible for the differences observed. The target gene(s) and the level of expression7–9, the concentration of double-stranded RNA (dsRNA) used10, combinations of different dsRNAs11, and delivery methods12 could affect RNAi efficacy. dsRNAs are delivered through transgenic plants or by topical applications including trunk injection, root absorption, or spraying2. Since deployment of genetically engineered trees in reforestation remains contentious5,13, non-transgenic RNAi methods may be more readily accepted by foresters, land managers, conservationists, and the general public.
RNAi works well in beetles14,15 and therefore, coleopteran forest pests including the emerald ash borer (Agrilus planipennis, EAB) and Asian longhorned beetle (Anoplophora glabripennis, ALB) are potential targets for RNAi-mediated control. Both beetles are non-native introductions that have become invasive in North America due to an abundance of highly susceptible host plant material and a lack of effective population regulators16,17. Both EAB and ALB have been shown to respond to RNAi, therefore this technology may be feasible for their control11,18.
Initial work with EAB demonstrated that oral delivery of dsRNA targeting iap (inhibitor of apoptosis) and cop (COPI coatomer, β subunit) genes causes mortality and gene silencing in neonate larvae after 10 days of exposure11. However, selection of optimal target gene(s) for a given insect requires extensive screening of multiple dsRNAs, which is typically done via micro-injection. In Tribolium castaneum, for instance, a large-scale screen identified 40 most effective genes that caused 50–100% mortality at 8 days post-injection9; eleven of those genes caused >80% mortality on day 6, and 100% on day 8 after injection.
Screening target genes for RNAi by microinjection of dsRNA is not an option for EAB neonate larvae because of their small size, delicate form, and their endophagous feeding habits11. Additionally, RNAi efficiency varies with delivery method, and dsRNA injection is typically more efficient than dsRNA feeding19. Finally, oral dsRNA delivery is essential given that one of our goals is to develop a method for controlling EAB using RNAi technology.
We first screened 13 genes by feeding dsRNA to neonate larvae and selected two target genes that caused the highest mortality. The expression of two selected genes was evaluated in EAB eggs, larvae, and adults, and knockdown of target genes after dsRNA exposure was determined in larvae and adults. Additionally, we evaluated the mortality rate after ingestion of individual and combined dsRNAs in both larvae and adults. Finally, a non-target organism, T. castaneum, was used to evaluate the specificity of the EAB.dsRNA we designed.
Results
Screening of candidate genes
The mortality of neonate larvae following ingestion of dsRNA varied among the genes tested with most causing 13 to 38% mortality (Fig. 1). After 4 d of dsRNA exposure, 13.2% mortality was recorded in negative controls (dsGFP and dsMalE), and 38% mortality in our positive control (dsIAP). However, ingestion of dsRNA targeting shi and hsp caused 55.5 and 63% mortality after 4 d, and reached 80 and 93.3% mortality, respectively, after 8 d of dsRNA exposure. Among the 13 genes targeted, ingestion of dsSHI and dsHSP was most effective in EAB neonates, causing the highest mortality following dsRNA exposure.
Figure 1.
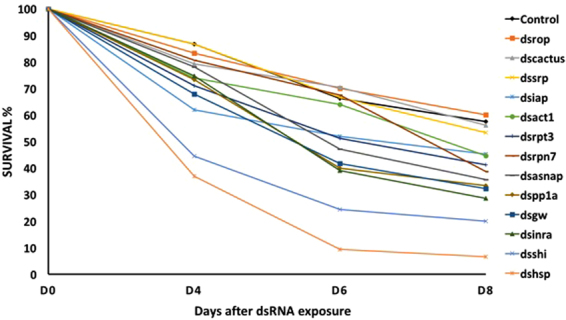
Screening of dsRNAs in EAB neonate larvae. Using a droplet feeding bioassay, neonate larvae were fed on dsRNAs at 10 µg/µL concentration for 4 d, followed by feeding sucrose solution without dsRNA. After day 4, mortality was recorded every 2 d until day 8. dsIAP was used as a positive control and dsmalE or dsGFP were negative controls. Bioassays were repeated twice for each treatment and 6x for control.
dsRNA dosage response
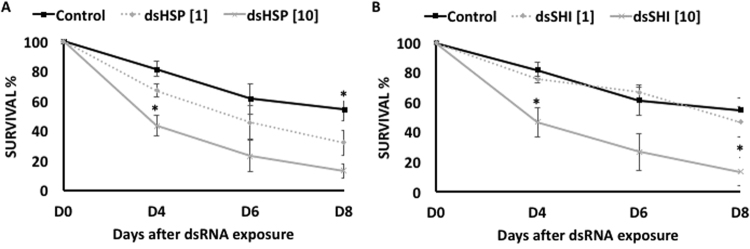
The two genes that caused the highest mortality, hsp and shi, were selected for further experiments to determine optimum dsRNA doses. Neonate larvae were fed on high (10 μg/μL of dsHSP or dsSHI) and low (1 μg/μL of dsHSP or dsSHI) doses of dsRNA. Neonates that ingested the high dose of either dsRNA experienced 90% mortality, whereas those that ingested low doses of dsHSP experienced 67% mortality after 8 d (Fig. 2A). In contrast, mortality of neonates ingesting 1 μg/μL of dsSHI did not differ from the control (Fig. 2B).
Figure 2.
EAB survival is evaluated after 8 d of feeding on dsRNA. (A) Neonate larvae were fed on two different concentrations of dsHSP for 4 days. Control = dsmalE or dsGFP at a concentration of 10 µg/µL (N = 6); dsHSP [1] = 1 µg/µL (N = 4); dsHSP [10] = 10 µg/µL (N = 3). (B) Neonate larvae were fed on two different concentrations of dsSHI for 4 days. dsmalE or dsGFP at a concentration of 10 µg/µL (N = 6) were used as control. dsSHI [1] = 1 µg/µL (N = 3); dsSHI [10] = 10 µg/µL (N = 4). The asterisk reflects significantly different mortality rate (ANOVA, Student-Newman-Keuls, P = 0.002 (A), P = 0.01 (B)).
Gene expression
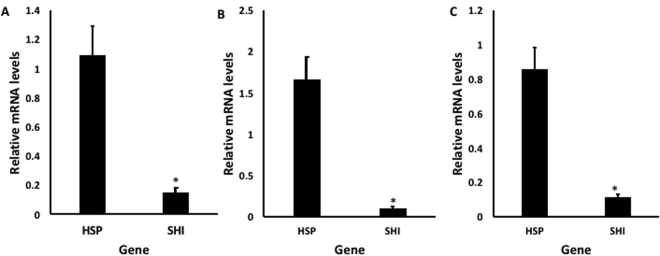
The relative expressions of hsp and shi genes were compared in EAB eggs, larvae, and adults (Fig. 3A–C). In all life stages tested, hsp gene expression is >8x more than shi gene expression.
Figure 3.
Relative expression of hsp and shi genes in EAB eggs, larvae, and adults. Eggs (A), neonate larvae (B) and adults (C) were exposed to dsGFP (control) and total RNA isolated after 48 h (A), 72 h (B), and 24 h (C). Relative mRNA levels were normalized using TEF as a reference gene. Mean + S.E (A: N = 3, B: N = 3; C: N = 5) are shown. The asterisk denotes significant differences (t-test, two-tailed P-value: P = 0.002 (A), P = 0.009 (B), P = 0.002 (C)).
Combination of dsRNA
Neonate larvae fed on 1 µg/µL of both dsHSP and dsSHI (500 ng/µL of each) for 4 d had higher mortality after 4, 6, and 8 d (61.6%, 73.8%, and 80.5%, respectively; Fig. 4) than the larvae fed on 1 µg/µL of dsHSP (Fig. 2A) or 1 µg/µL of dsSHI (Fig. 2B).
Figure 4.
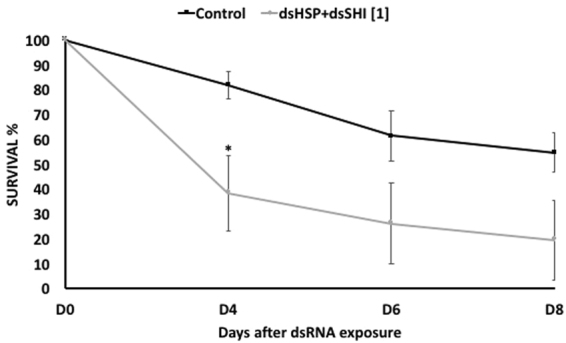
Larval EAB survival after 8 days fed on a combination of dsRNAs. Neonates were exposed to 1 µg/µL of dsRNA (500 ng/µL each dsHSP and dsSHI; N = 3) for 4 consecutive days and then fed on blue-sucrose solution without dsRNA until day 8; 10 µg/µL of dsmalE or dsGFP (N = 6) were used as control. The asterisk denotes days of exposure when treatments are significantly different (t-test, two-tailed P-value: P = 0.007).
Gene silencing
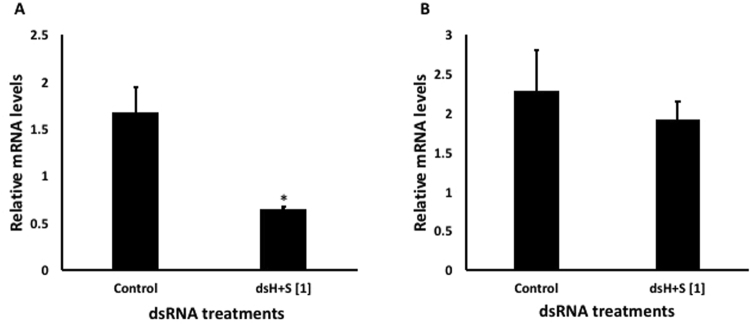
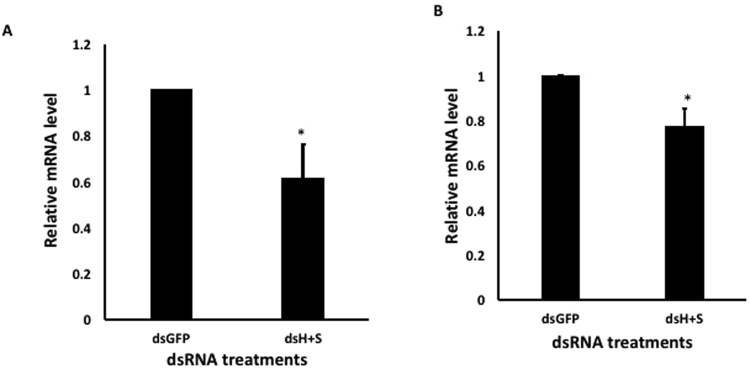
Neonates fed on 1 µg/µL of both dsHSP and dsSHI (500 ng/µL of each) for 3 d showed a decrease in the mRNA levels of hsp (62%) and shi (15%) when compared to their levels in neonates that fed on dsGFP (Fig. 5A,B). In adults, injection of 20 µg/µL of both dsHSP and dsSHI (10 µg/µL of each) resulted in 20–40% silencing (p ≤ 0.05) of both hsp and shi genes at 24 h after injection (Fig. 6A,B).
Figure 5.
Relative expression of hsp (A) and shi (B) genes in EAB larvae after 3 days feeding on 1 µg/µL of combined dsRNAs (500 ng/µL each dsHSP and dsSHI); 10 µg/µL of dsGFP was used as control. Relative mRNA levels were normalized using TEF as a reference gene. Mean + S.E (N = 3) are shown. The asterisk above the bar indicates significantly different expression (t-test, two-tailed P-value: P = 0.038 (A), P = 0.635 (B)).
Figure 6.
Relative expression of hsp (A) and shi (B) genes in EAB adults 24 h after injection of 20 µg/µL of dsRNA (10 µg/µL each dsHSP and dsSHI); 20 µg/µL of dsGFP were used as control. Relative mRNA levels were normalized using TEF as a reference gene. Mean + S.E (N = 5) are shown (t-test, two-tailed P-value: P = 0.05 (A), P = 0.02 (B)).
Adult mortality
In the adult feeding trial, beetles were fed on 10 µg/µL dsSHI, 10 µg/µL dsHSP, 1 µg/µL dsHSP + dsSHI (500 ng/µL of each), or a 10 µg/µL dsGFP control. There were no effects detected on day 1 with the exception of a single beetle lost in the control treatment, which then remained constant through day 13. Adult beetles fed on dsSHI experienced 30% mortality at 6 d, which persisted through the experiment. Adult mortality became evident at 3 d in adults fed on dsHSP (10%); mortality rose to 40% for dsHSP-exposed beetles for the duration of the experiment. However, for beetles fed on 1µg/µL of combined dsHSP and dsSHI, 90% mortality was evident on day 13 (Fig. 7).
Figure 7.
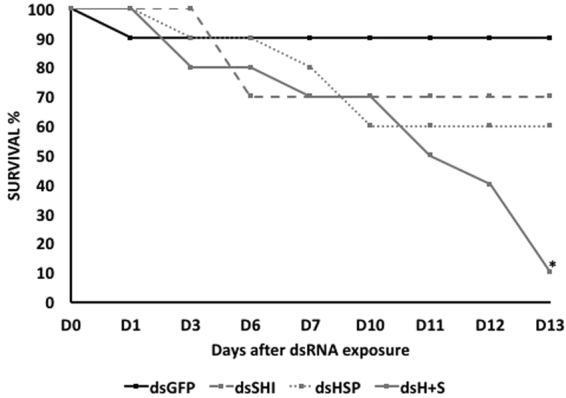
Adult EAB survival at 14 days after feeding on 2 µL of dsRNA at a final concentration of 10 µg/µL of single dsGFP (control), single dsHSP, and single dsSHI and 1 µg/µL of combined dsHSP and dsSHI (500 ng/µL of each). The asterisk indicates significant differences in mortality between treatment and control (Fisher’s Exact test, two-tailed, P-value = 0.011, N = 10).
EAB.dsRNA in Tribolium castaneum
The nucleic acid sequences of the EAB hsp and shi showed 85% and 91% identity, respectively, with the homologues in T. castaneum. Injection of EAB dsHSP and dsSHI into T. castaneum adults caused no mortality (Table 1).
Table 1.
Similarity between A. planipennis dsRNA designed sequences and T. castaneum genes.
| Gene Name |
T. castaneum Accession |
Query Cover |
E-value | Identity | Mortality* (10 dai) |
|---|---|---|---|---|---|
| shi | XM_008200600.2 | 92% | 3e-49 | 91% | 0% |
| hsp | XM_015982882.1 | 78% | 7e-29 | 85% | 0% |
dai: days after dsRNA injection. *Sun-Shepard’s formula was used to adjust for mortality.
Discussion
Adoption of RNAi into management strategies against forest pests lags behind that of agricultural pests, but numerous native and non-native forest insect pests are appropriate candidates for this technology. Emerald ash borer is a non-native, phloem-feeding beetle that has killed millions of trees in North America; its invasion is unprecedented in scope and magnitude17. Although chemical suppression is possible in managed settings20 and classical biological control has been implemented with considerable success21,22, the invaded range continues to expand and ash trees continue to die. The need for innovative approaches to EAB management remains high17.
We are investigating the feasilibility of utilizing RNAi as a means of suppressing EAB, and previously demonstrated that EAB has efficient RNAi machinery, and that neonates ingesting dsIAP and dsCOP, targeting iap and cop genes, experienced significant mortality11. However, selection of target genes with rapid action is essential, as is an oral method to deliver dsRNA molecules to EAB. Here, we used a neonate larval feeding assay to screen 13 target genes, and selected two that are especially efficacious and demonstrate potential for incorporation into an EAB management scheme.
A significant mortality was observed 4 to 8 days after dsRNA ingestion, depending on dsRNA concentration and target gene, corroborating data reported previously in T. castaneum9. At 8 d after dsRNA ingestion, the effect of the dsRNAs causing EAB neonate mortality clustered into three groups (Fig. 1). The first group has three genes that caused 40–47% mortality and did not differ from the effects observed in the control treatment (45%). The second group of eight genes caused 55–72% mortality and included the positive control dsIAP. However, the third group included the genes shi and hsp, which caused 80% and 93% mortality, respectively. Both shi and hsp caused significant mortality after 4 and 6 d following ingestion of dsRNAs targeting these specific genes.
We then evaluated the effects of these selected dsRNAs in two different concentrations. In studies using Bactrocera dorsalis (Diptera: Tephritidae), the concentration of dsRNA influenced the level of gene suppression23, and in studies using oral delivery of dsRNA to Diabrotica v. virgifera and A. planipennis, there was a positive correlation between dsRNA concentration and mortality rate11,24. The most efficient mortality response in EAB was observed on the 8th day after ingestion of the highest concentration of dsHSP and dsSHI (~90%; Fig. 2A,B). Ingestion of the lower concentration of dsHSP alone also caused a significant larval mortality (67%), but the lower concentration of dsSHI alone (53% mortality) did not differ from the control (45%). One explanation for differences in efficiency among target genes may be attributed to the relative expression profile of the target genes. In EAB eggs, neonate larvae, and adults, the hsp gene is highly expressed relative to shi (Fig. 3). Highly expressed genes are considered more susceptible to RNAi gene silencing responses, suggesting that the abundance of target mRNAs is one of the critical factors that determine the efficiency of the RNAi in a given cellular context7. Corroborating this, we also found that the relative silencing of hsp was greater than shi silencing (62% versus 15%) (Fig. 5). It is interesting that ingestion of a combination of two dsRNAs at low concentrations (500 ng of each dsSHI and dsHSP) by neonates and adults efficiently induced mortality. Exposing neonates to dsSHI + dsHSP, in a final concentration of 1 μg/μL caused 62% mortality in 4 days (Fig. 4), whereas, dsHSP alone caused 33%, and dsSHI alone caused 25% mortality (Fig. 2A,B, respectively).
Even larvae that were exposed to a 10x higher concentration of single dsRNA treatment showed lower mortality than that observed after treatment with both the dsRNAs, and similar results were observed in adults. After 2 weeks, adults fed with combined dsRNA in 10x lower concentration experienced 90% mortality (Fig. 7). On the other hand, when the insects were fed on a single dsRNA in high concentration, 30–40% of mortality was observed. Although competition among multiple dsRNA’s can potentially confound RNAi efficiency25, our findings corroborate previous studies that showed additive effects of combined dsRNA9,11 in EAB. As a target, shi appears to be less efficient, but when supplemented with hsp, shi may become more potent. However, further studies on gene expression patterns and other combinations of genes must be performed to better understand and confirm these findings.
These two genes, shi and hsp, were silenced after dsRNA injection in T. castaneum adults, where they caused 100% of mortality9. In our study, after injecting T. castaneum with dsHSP and dsSHI designed specifically to silence EAB genes, no mortality was observed after 10 days. Our results suggest that EAB.dsSHI and EAB.dsHSP appear specific to our target insect and appear to have negligible effects in a non-target organism. However, bioassays must be performed to confirm the RNAi effects in other non-target organisms, especially the classical biological control agents Tetrastichus planipennisi (Eulophidae), Spathius agrili and S. galinae (Braconidae), and Oobius agrili (Encyrtidae), that form the mainstay of sustainable EAB management programs in North America21. Studies must also be performed to evaluate its safety for other off-target organisms, as well as dsRNA stability, transport inside the plant, and its fate in the environment1. Finally, development of applied systems to deliver dsRNAs remains to be developed. Encapsulated dsRNA in nanoparticles to be used as a foliar and trunk spray, soil treatment and root absorption, and tree trunk injections are some of the topical applications; genetically engineered trees expressing dsRNA are another possibility2,26,27.
Our findings suggest that hsp and shi are potential target genes to suppress EAB populations. As EAB continues its invasion of North America, we continue to accumulate additional non-native species, and endemic pests undergo extensive and destructive population fluctuations, we must increase our efforts to develop innovative approaches for forest pest management. Our work demonstrates that RNAi may become a feasible approach to suppressing pests in forests.
Methods
Insects
Laboratory-reared A. planipennis eggs were placed in Petri dishes (150 × 15 mm) with moistened filter paper and maintained at 23 °C and 75% relative humidity in a growth chamber. Newly hatched unfed neonate larvae at <48 h post-hatch were used in bioassays. Adults were reared in the laboratory at 23 °C from field-collected green ash (F. pennsylvanica), and maintained on either green or tropical ash (F. uhdei) foliage.
Selection of target genes and synthesis of dsRNA
PCR templates for in vitro transcription of dsRNA were generated using gene-specific primers containing T7 polymerase promoter sequence (TAATACGACTCACTATAGGG) at the 5’end (Table 2). Candidate genes were selected based on previous reports of mortality in T. castaneum exposed to dsRNAs targeting these genes9. PCR conditions were 94 °C for 4 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 45 s, finishing with an extension step at 72 °C for 10 min. The PCR template was purified using a PCR purification kit (Qiagen Inc., Valencia, CA USA). As a negative control, a fragment of GFP (green fluorescence protein) was used. After PCR purification, dsRNA synthesis was performed using the MEGAscript RNAi Kit (Ambion Inc., Foster City, CA USA) following manufacturer’s instructions. Briefly, 200 ng of purified PCR product was used as template in a 20 μL in vitro transcription reaction. The reaction was incubated for 16 h at 37 °C, followed by 15 min of DNase treatment. The dsRNA was precipitated by adding 0.1x volume of sodium acetate (3 M, pH 5.2) and 2.5x the volume of 100% ethanol; kept at −20 °C for at least 2 h followed by centrifugation at 4 °C for 30 min. The dsRNA pellet was then rinsed with 750 μL of 75% ethanol and centrifuged again at 4 °C for 15 min. The ethanol was removed and the dsRNA was diluted in ultrapure distilled water. The quality of the dsRNA was checked by electrophoresis and quantified using a spectrophotometer (NanoDrop Technologies, Wilmington, DE USA). When a higher concentration of dsRNA was needed, the samples were vacuum concentrated using Concentrator plus (Eppendorf, Hauppauge, NY, USA).
Table 2.
Primer sequence, dsRNA size and NCBI sequence ID for the target genes. All primers contain the promoter sequence of T7 RNA Polymerase that is not represented in the table.
| Gene Name | Primer name-dsRNA size | Primer Sequence | Ap sequence ID |
|---|---|---|---|
| srp54k | ds - EAB srp - 496 bp – F | GGCTGTAGCAAATGCAGTGA | XM_018478973.1 |
| ds - EAB srp - 496 bp - R | ACGCGCCATAGACTCTTGTT | ||
| ras opposite | ds - EAB rop - 493 bp – F | GCCGACACCTTTCAGTGTTT | XM_018464595.1 |
| ds - EAB rop - 493 bp - R | GCTTGGAATCGGTGAACTTT | ||
| shibire | ds - EAB shi -483 bp – F | TGGCACATTTGTATGCCAGT | XM_018465318.1 |
| ds - EAB shi -483 bp - R | CTTGTTGCATTTGCTGAGGA | ||
| protein phosphatase 1 alpha at 96a | ds - EAB pp1a - 532 bp – F | TATGTGTACGTGTGCCCGTT | XM_018468028.1 |
| ds - EAB pp1a - 532 bp - R | TTGATGAAGAGCAAGCGAAA | ||
| regulatory particle non-atpase 7 | ds - EAB rpn7 - bp – F | TTGAAGAGGGAGGTGATTGG | XM_018468408.1 |
| ds - EAB rpn7 - bp - R | TGATCCGGCCTATTTGTCTC | ||
| regulatory particle triple-a atpase 3 | ds - EAB rpt3 - 513 bp – F | GCCAAAGCAGTAGCACATCA | XM_018479000.1 |
| ds - EAB rpt3 - 513 bp - R | AGCATGCATTCCAGCTTCTT | ||
| actin | ds - EAB act1 - 427 bp – F | GCTAACCGCGAGAAGATGAC | XM_018481271.1 |
| ds - EAB act1 - 427 bp - R | GGAACCTTTCGTTTCCAACA | ||
| cactus | ds - EAB cact - 389 bp – F | ATGTTGTGTTGGTGCGAAAA | XM_018478973.1 |
| ds - EAB cact - 389 bp - R | TTCCCGAACTAAGGTCGTTG | ||
| alpha snap | ds - EAB asnap - 484 bp – F | GTAGTGCATTTTGCGAAGCA | XM_018465131.1 |
| ds - EAB asnap - 484 bp - R | TGACGAGCATTCAGCAAATC | ||
| inverse regulator a | ds - EAB inra - 449 bp – F | ACCGGTGTTACCAAAGCAAG | XM_018464445.1 |
| ds - EAB inra - 449 bp - R | GCGCTATTAACAGGCGCTAC | ||
| heat shock 70-kDa protein cognate 3 | ds - EAB hsc70-3 - 468 bp – F | GTTACGAGCCAGGGTGAAAA | XM_018474521.1 |
| ds - EAB hsc70-3 - 468 bp - R | CTTTTGAACGGCACGGTTAT | ||
| gawky | ds - EAB gw - 442 bp - F | CAACATTGCGCCGACTACTA | XM_018475118.1 |
| ds - EAB gw - 442 bp - R | CCACATTCCTCCTCCACTGT |
Screening assays
A droplet bioassay11 was used to screen potential target genes for EAB suppression. Twelve candidate genes (Table 2) and positive and negative controls were evaluated. Inhibitor of apoptosis gene (iap 1), previously shown to cause mortality after dsIAP exposure in both EAB and ALB11,18, was used as a positive control, and green fluorescent protein (gfp) was used as a negative control. Droplets of dsRNA (2 μl each) at a concentration of 10 μg/μl diluted in blue sucrose solution were used to feed 3–4 neonate EAB larvae in covered petri dishes with moistened filter paper. The blue dye in the sucrose-dsRNA solution allowed us to confirm neonate larval consumption. Neonates were exposed to dsRNA for 4 consecutive days; droplets were replenished on the rare occasions when the solution was completely consumed or evaporated. On day 5, larvae were transferred to new plates containing droplets of sucrose solution without dsRNA. Larval mortality was recorded at 48 h intervals through day 8 after dsRNA feeding treatment. Experiments were repeated twice under the same conditions using 20 neonate larvae per treatment. If the two repeats showed variability, a third repeat was performed. The dsRNAs that showed higher mortality after 8 d were repeated at least one more time, for a total of three repeats; those with the highest EAB mortality were selected for further experiments to confirm their potential for use in managing EAB.
Dosage response
Two concentrations, 1 μg/μL and 10 μg/μL, of dsHSP (targeting heat shock 70-kDa protein cognate 3 gene) and dsSHI (targeting shibire gene) were used to evaluate neonate mortality, and 10 μg/μL dsGFP was used as a control. Larvae were fed on dsRNA for 4 d, followed by 4 d on blue sucrose solution without dsRNA. Mortality (%) was then calculated based on the initial number of larvae (15–20) on day one. The experiment was repeated 3–6 times, and a one-way analysis of variance (ANOVA) was used with Student-Newman-Keuls to evaluate significance of differences.
Combinations of dsRNA
The two selected dsRNAs, dsHSP and dsSHI, were combined to evaluate larval mortality and gene silencing using the droplet assay. The dsRNAs were diluted in sucrose solution to a final concentration of 10x lower single dsRNA (1 μg/μL dsSHI + dsHSP: 0.5 μg/μL of dsSHI and 0.5 μg/μL of dsHSP). The dsGFP at a concentration of 10 μg/μL was used as a control. Four biological replicates, with 15–20 neonate larvae per replicate, were used and a one-way ANOVA was used for data analysis, with Student-Newman-Keuls to detect significance of differences.
Gene expression
Both hsp and shi genes were analyzed for gene expression patterns in eggs, larvae, and adults, and for knockdown after dsRNA exposure in neonate larvae and adults, using dsGFP as a control. For gene expression patterns, the control treatment in neonate larvae and adults was used for the analyses. The eggs were treated with 1 μg/μL of dsGFP followed by a 1 μL droplet of dsRNA (3 times during 48 h). Eggs were maintained at 28 °C for 48 h, and each treatment was performed in triplicate with 6–7 eggs per replicate. For gene knockdown analyses, neonate larvae were fed on both dsSHI and dsHSP (1 μg/μL total, 0.5 μg/μL of each dsRNA) for 72 h; 10 μg/μL of dsGFP was used as a control. Larval assays were performed in triplicate with 6–8 neonates per replicate. Adult beetles were evaluated for hsp and shi gene silencing 24 h after injection with 2 μL of both dsRNAs (dsSHI and dsHSP at 20 μg/insect total, 10 μg/μL of each dsRNA), using dsGFP (20 μg/insect) as a control. Micro-injection of dsRNA was chosen because it is faster, easier, and requires fewer insects to perform. Five biological replicates were performed using one beetle in each replicate. Following assays, eggs and neonate larvae were pooled for RNA extraction. RNA was also isolated from each adult beetle. A two-tailed t-test was used for statistical analysis to compare the means of a single variable.
RT-qPCR
Total RNA was isolated from 6–10 EAB larvae pooled after feeding on dsRNA using the TRI Reagent RT (Molecular Research Center Inc., Cincinnati, OH, USA). The cDNA was synthesized using M-MLV Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA) from RNA and used as a template for gene expression studies. The expression analyses of the target genes were conducted using SYBR Green PCR Master Mix. Briefly, the PCR mixture contained 1 μL of synthesized cDNA, 0.2 μL of each primer (10 mM; Table 3), 5 μL of the SYBR green PCR master mix and 3.6 μL of ddH2O. The reactions were carried out in duplicate per template in a final volume of 10 μL. RT-qPCR reactions were performed by the StepOnePlus Real-Time PCR system (Life Technologies, Carlsbad, CA, USA) using the following cycling conditions: one cycle at 95 °C (20 s), followed by 40 cycles of denaturation at 95 °C (3 s), annealing and extension at 60 °C for 30 s. At the end of each RT-qPCR reaction, a melting curve was generated to confirm a single peak and rule out the possibility of primer-dimer and non-specific product formation. The TEF1A was used as reference gene28, and 2−ΔΔCt method was used to calculate the relative expression levels of the target gene in the samples compared to controls29. Standard curves were performed for all the new primers and the correlation coefficients and amplification efficiency parameters were analyzed. A two-tailed t-test was used for statistical analysis to compare the means of a single variable.
Table 3.
Primer sequence and amplicon size for the target genes shi and hsp, for qPCR.
| Primer Name | Primer Sequence (5′- 3′) | Amplicon (bp) |
|---|---|---|
| q- EAB shi - 122 bp - F | GGGATCTGCCCAAATTAACA | 122 |
| q - EAB shi - 122 bp - R | CCCGTCTGAGTTCTTTCTCG | |
| q- EAB hsp70-3 - 97 bp - F | GACAAAGGAACGGGAAACAA | 97 |
| q - EAB hsp70-3 - 97 bp - R | TCTCGGCATCCCTTATCATC |
Adult mortality
A feeding assay was performed to deliver dsRNAs to EAB adults to evaluate the effects of silencing hsp and shi genes. The dsRNAs were diluted with 40% sucrose solution, and each beetle was fed on ~2 µL droplet of the diluted solution of each dsRNA sample. Beetles were fed 10 µL of each dsRNA (dsGFP, dsHSP, and dsSHI), and 1 µL of the combination of dsHSP and dsSHI (500 ng of each). In each treatment, 5 male and 5 female beetles (N = 10) were evaluated. Mortality was recorded at 24–48 h intervals for 13 d. After the single dsRNA feeding exposure at day 0, adult beetles were provided with fresh tropical ash foliage and were kept in an environmental chamber (Percival Scientific, Perry, IA) at 25 + 1.5 °C, 55–65% RH, and a photoperiod of 16:8 (L:D) h. For data analysis, males and females were combined, and Fisher’s test was used to identify statistical differences between treatments.
EAB dsRNA effect on Tribolium castaneum
To evaluate potential off-target effects of the EAB-specific dsRNA, both EAB.dsHSP and EAB.dsSHI or dsGFP as a control were injected into T. castaneum adults. For microinjections, 10–18 adults were anesthetized on ice and temporarily fixed on a glass slide using double-sided tape. 0.1 μL of 5 μg/μL dsRNA (0.5 μg/adult) was injected into the dorsal side of its first or second abdominal segment using Nanoject III Programmable Nanoliter Injector (Drummond Scientific Company, Broomall, PA, USA). After dsRNA injections insects were place in 12-well plates with wheat flour and held in darkness at 26 °C and 36% humidity. Insect mortality was recorded 10 d post-dsRNA injection. Sun-Shepard’s formula30 was used to adjust for mortality of controls. Bioinformatic analyses were performed to compare the sequences of gene fragment used for dsRNA synthesis from EAB and T. castaneum. Sequences of hsp and shi gene regions used for dsRNA synthesis from EAB were aligned to T. castaneum genes using MUSCLE software.
Acknowledgements
This is publication number 17–08–109 from the Kentucky Agricultural Experimental Station and is published with the approval of the director. This work was supported by the University of Kentucky, the USDA Forest Service Forest Health Research and Education Center, and the Kentucky Agricultural Experiment Station under McIntire-Stennis 2351197000.
Author Contributions
T.B.R., L.K.R., and S.R.P. conceived the experiments; J.J.D. provided insects and conducted the adult assay; T.B.R. conducted the experiments; T.B.R., L.K.R., J.J.D., and S.R.P. analyzed the results; T.B.R., L.K.R., J.J.D., and S.R.P. prepared the manuscript. All authors approved the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bachman PM, et al. Ecological risk assessment for DvSnf7 RNA: A plant-incorporated protectant with targeted activity against western corn rootworm. Regul Toxicol Pharmacol. 2016;81:77–88. doi: 10.1016/j.yrtph.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Joga MR, Zotti MJ, Smagghe G, Christiaens O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Frontiers in Physiology. 2016;7:553. doi: 10.3389/fphys.2016.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burand JP, Hunter WB. RNAi: Future in insect management. J Invertebr Pathol. 2013;112:S68–S74. doi: 10.1016/j.jip.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Palli SR. RNAi methods for management of insects and their pathogens. DNA. 2012;5:3. [Google Scholar]
- 5.Gamborg, C. & Sandøe, P. Ethical considerations regarding genetically modified trees. IUFRO Forests and Genetically Modified Trees. Food and Agriculture Organization of the United Nations, Rome, 163–176 (2010).
- 6.Fladung, M., Pasonen, H. & Walter, C. Genetically modified trees and associated environmental concerns. IUFRO forests and genetically modified trees. Food and Agriculture Organization of the United Nations, Rome, 177–202 (2010).
- 7.Hong SW, Jiang Y, Kim S, Li CJ, Lee D-K. Target Gene Abundance Contributes to the Efficiency of siRNA-Mediated Gene Silencing. Nucleic Acid Therapeutics. 2014;24:192–198. doi: 10.1089/nat.2013.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 9.Ulrich, J. et al. Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics16, 10.1186/s12864-015-1880-y (2015). [DOI] [PMC free article] [PubMed]
- 10.Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol. 2009;39:824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues TB, et al. S. R. Development of RNAi method for screening candidate genes to control emerald ash borer, Agrilus planipennis. Scientific Reports. 2017;7:7379. doi: 10.1038/s41598-017-07605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott JG, et al. Towards the elements of successful insect RNAi. J Insect Physiol. 2013;59:1212–1221. doi: 10.1016/j.jinsphys.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häggman H, et al. Genetically engineered trees for plantation forests: key considerations for environmental risk assessment. Plant Biotechnol J. 2013;11:785–798. doi: 10.1111/pbi.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu F, Xu JJ, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle. Leptinotarsa decemlineata. Pest Manage Sci. 2011;67:175–182. doi: 10.1002/ps.2048. [DOI] [PubMed] [Google Scholar]
- 15.Palli SR. RNA interference in colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Current Opinion in Insect Science. 2014;6:1–8. doi: 10.1016/j.cois.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Angeli S, Schuetz S, Luo Y, Hajek AE. Ecology and management of exotic and endemic Asian longhorned beetle Anoplophora glabripennis. Agricultural and Forest Entomology. 2009;11:359–375. doi: 10.1111/j.1461-9563.2009.00443.x. [DOI] [Google Scholar]
- 17.Herms DA, McCullough DG. Emerald ash borer invasion of North America: history, biology, ecology, impacts, and management. Annu Rev Entomol. 2014;59:13–30. doi: 10.1146/annurev-ento-011613-162051. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. RNA interference in the Asian Longhorned Beetle:Identification of Key RNAi Genes and Reference Genes for RT-qPCR. Scientific Reports. 2017;7:8913. doi: 10.1038/s41598-017-08813-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, et al. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem Mol Biol. 2016;77:1–9. doi: 10.1016/j.ibmb.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Herms, D. A. et al. Insecticide options for protecting ash trees from emerald ash borer. North central IPM center bulletin12 (2009).
- 21.Duan JJ, Abell KJ, Bauer LS, Gould J, Van Driesche R. Natural enemies implicated in the regulation of an invasive pest: a life table analysis of the population dynamics of the emerald ash borer. Agricultural and Forest Entomology. 2014;16:406–416. doi: 10.1111/afe.12070. [DOI] [Google Scholar]
- 22.Davidson W, Rieske LK. Establishment of classical biological control targeting emerald ash borer is facilitated by use of insecticides, with little effect on native arthropod communities. Biological Control. 2016;101:78–86. doi: 10.1016/j.biocontrol.2016.06.010. [DOI] [Google Scholar]
- 23.Li, X. X., Dong, X. L., Zou, C. & Zhang, H. Y. Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Scientific Reports5, 10.1038/srep08700 (2015). [DOI] [PMC free article] [PubMed]
- 24.Galdeano DM, Breton MC, Lopes JRS, Falk BW, Machado MA. Oral delivery of double-stranded RNAs induces mortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri. Plos One. 2017;12:e0171847. doi: 10.1371/journal.pone.0171847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller SC, Miyata K, Brown SJ, Tomoyasu Y. Dissecting Systemic RNA Interference in the Red Flour Beetle Tribolium castaneum: Parameters Affecting the Efficiency of RNAi. Plos One. 2012;7:e47431. doi: 10.1371/journal.pone.0047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taning, C. N. T., Andrade, E. C., Hunter, W. B., Christiaens, O. & Smagghe, G. Asian Citrus Psyllid RNAi Pathway - RNAi evidence. Scientific Reports6, 10.1038/srep38082 (2016). [DOI] [PMC free article] [PubMed]
- 27.Andrade EC, Hunter WB. RNAi feeding bioassay: development of a non-transgenic approach to control Asian citrus psyllid and other hemipterans. Entomol Exp Appl. 2017;162:389–396. doi: 10.1111/eea.12544. [DOI] [Google Scholar]
- 28.Rajarapu SP, Mamidala P, Mittapalli O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis) Insect Sci. 2012;19:41–46. doi: 10.1111/j.1744-7917.2011.01447.x. [DOI] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using RealTime Quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Puntener, W. Manual for field trials in plant protection. Second Edition edn (Ciba-Geigy Limited, 1981).