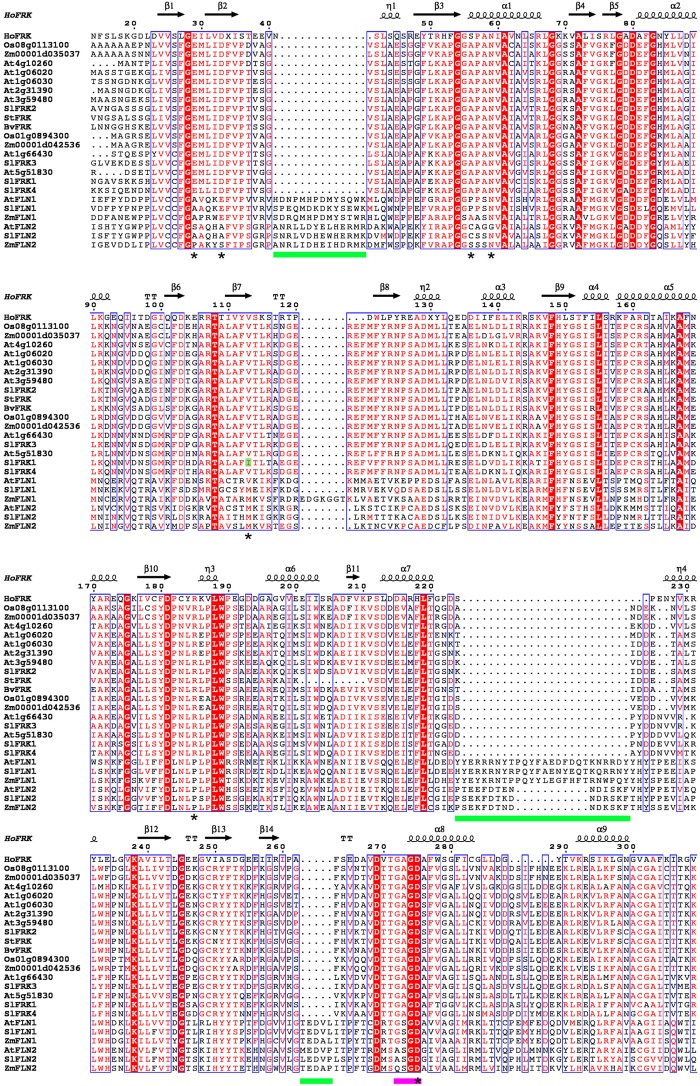
FIGURE 2.
Sequence alignment of plant FRKs and FLNs. Sequence alignment was carried out using ClustalW and the figure was created using ESPript 3.0 (Robert and Gouet, 2014). The structure of H. orenii FRK (PDB – 3HJ6) was used as a reference structure. N-terminal and C-terminal regions were excluded from the graphic presentation. A red background indicates amino acid sequence identity and red letters indicate sequence similarity. Green bars show the locations of insertions in FLNs. A pink bar is used to indicate the G/AXGD motif. Asterisks indicate suggested amino acids with importance for substrate binding (Chua et al., 2010). Accession IDs of the proteins used for the sequence alignment: SlFRK1 (AAB57733), SlFRK2 (AAB57734), SlFRK3 (NP_001234396), SlFRK4 (AAM44084), At1g06020 (AAF80125), At1g06030 (AAF80126), At1g66430 (AAG51160), At2g31390 (AAM14251), At3g59480 (CAB75445), At4g10260 (CAB78149), At5g51830 (AAL34211), Os01g0894300(AAL26574), Os08g0113100 (AAL26573), Zm00001d035037 (AAP42806), Zm00001d042536 (AAP42805), BvFRK (AAA80675), StFRK (CAA78283), AtFLN1 (AEE79187), AtFLN2 (NP_177080), SlFLN1 (XP_004246362), SlFLN2 (XP_004239035), ZmFLN1 (ONM36391),and ZmFLN2 (ONM06924).