FIGURE 3.
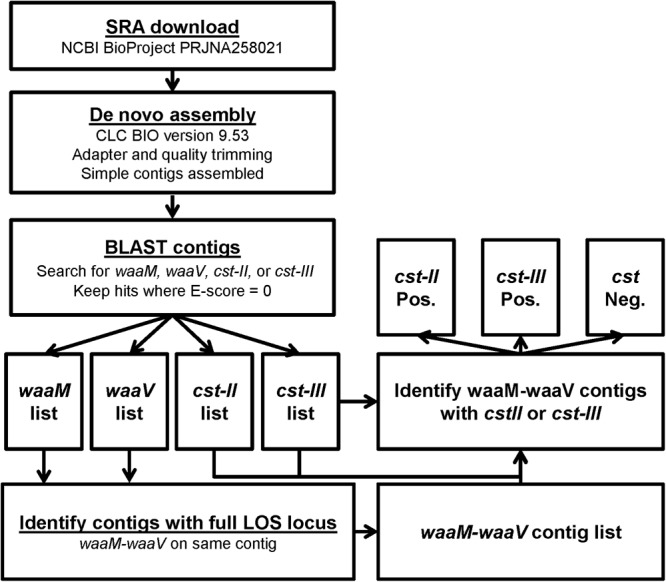
Workflow for the determination of cst-type from GenomeTrakr SRA data. The strategy for identifying cst genes from SRA files starts with de novo assembly, followed by BLAST searches for the four genes of interest. To ensure that the variable genes of the LOS locus were present in an SRA, contigs were identified that contained both flanking genes, waaM and waaV. The waaM-waaV contigs were then determined to be cst-II positive or cst-III positive if the gene was found in the same contig, and cst negative if the genes were not detected.
