Fig. 2.
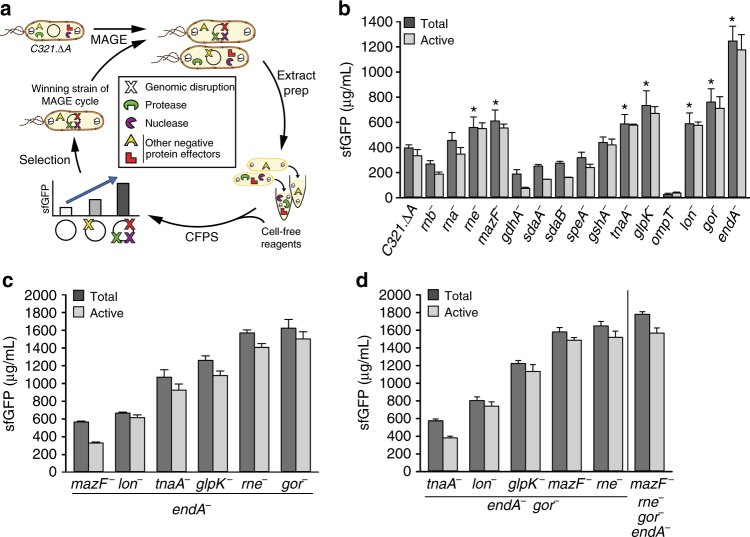
Engineering C321.∆A variants for enhanced CFPS. a Schematic of design-build-test cycles employing multiplex automated genome engineering (MAGE) to disrupt putative negative protein effectors (Supplementary Table 1) in engineered C321.∆A strains for producing extracts with enhanced cell-free protein synthesis (CFPS) yields. b Cell extracts derived from C321.∆A and genomically engineered strains containing a single putative negative effector inactivation were screened for sfGFP yields. Beneficial mutations that increase active yields ≥ 50% relative to C321.ΔA are highlighted with an *(p < 0.01, Student’s t-test). c C321.∆A.542 (endA−) was chosen as the next base strain and the following beneficial disruptions were pursued in combination: rne, mazF, tnaA, glpK, lon, and gor. d C321.∆A.709 (endA− gor−) was selected as the subsequent base strain for triple and quadruple mutant construction. C321.∆A.759 (endA− gor− rne− mazF−) yielded the highest level of CFPS production. Total sfGFP concentration was measured by counting radioactive 14C-Leucine incorporation and active protein was measured using fluorescence. Three independent batch CFPS reactions were performed for each sample at 30 °C for 20 h (n = 3). Error bar = 1 SD