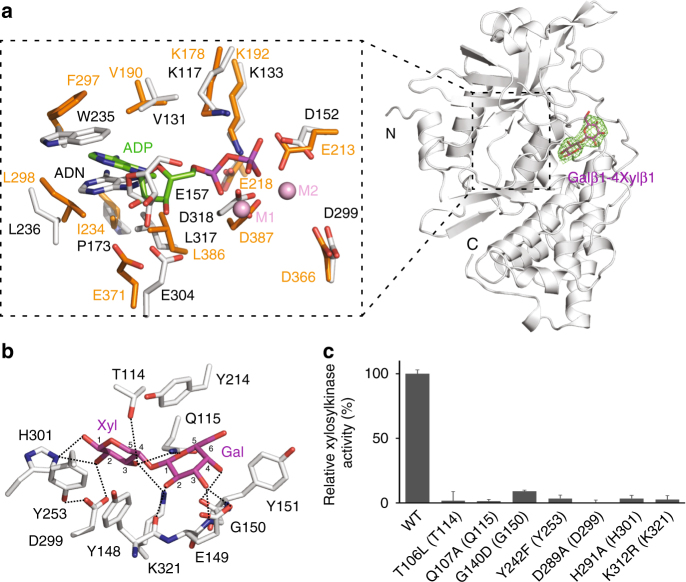
Fig. 6.
Substrate recognition mechanism of Fam20B. a The structure of hmFam20 in complex with Gal-Xyl. hmFam20 is shown as white ribbons. The Fo−Fc difference electron density map (3.0 σ) calculated before Gal-Xyl was modeled is shown as a green mesh, revealing the presence of the disaccharide. A close view of the ATP-binding site is displayed on the left, with the Mn/ADP-bound ceFam20 structure (shown in orange) superimposed for comparison. ADN, adenosine. The two Mn ions in ceFam20 are labeled as M1 and M2. b An enlarged image of the disaccharide-binding pocket showing the detailed molecular interactions between hmFam20 and Gal-Xyl. c Human Fam20B mutants display reduced or abolished kinase activity. The amino acids in brackets indicate the corresponding residues in hmFam20. Error bars represent the standard deviation of three independent experiments