Abstract
A subset of hematologic cancer patients is refractory to treatment or suffers relapse, due in part to minimal residual disease, whereby some cancer cells survive treatment. Cell-adhesion-mediated drug resistance is an important mechanism, whereby cancer cells receive survival signals via interaction with e.g. stromal cells. No genome-wide studies of in vitro systems have yet been performed to compare gene expression in different cell subsets within a co-culture and cells grown separately. Using RNA sequencing and species-specific read mapping, we compared transcript levels in human Jeko-1 mantle cell lymphoma cells stably adhered to mouse MS-5 stromal cells or in suspension within a co-culture or cultured separately as well as in stromal cells in co-culture or in separate culture. From 1050 differentially expressed transcripts in adherent mantle cell lymphoma cells, we identified 24 functional categories that together represent four main functional themes, anti-apoptosis, B-cell signaling, cell adhesion/migration and early mitosis. A comparison with previous mantle cell lymphoma and chronic lymphocytic leukemia studies, of gene expression differences between lymph node and blood, identified 116 genes that are differentially expressed in all three studies. From these genes, we suggest a core set of genes (CCL3, CCL4, DUSP4, ETV5, ICAM1, IL15RA, IL21R, IL4I1, MFSD2A, NFKB1, NFKBIE, SEMA7A, TMEM2) characteristic of cells undergoing cell-adhesion-mediated microenvironment signaling in mantle cell lymphoma/chronic lymphocytic leukemia. The model system developed and characterized here together with the core gene set will be useful for future studies of pathways that mediate increased cancer cell survival and drug resistance mechanisms.
Introduction
Novel therapy regimes have improved the prognosis for many types of lymphoma and leukemia.1 However, unsolved problems remain for many patients who are refractory to treatment or experience disease relapse. For example, mantle cell lymphoma (MCL) is an aggressive and generally incurable B-cell neoplasm, comprising about 8% of non-Hodgkin lymphomas (NHL).2 MCL is characterized by a high relapse rate and frequent resistance to therapy, which together with high median age at diagnosis makes MCL difficult to cure. While median overall survival time has doubled in recent years,3 the prognosis for MCL patients is poor with fewer than 15% long-term survivors.4
Signaling pathway defects intrinsic to MCL cells, due to genetic aberrations, provide only a partial explanation for its aggressiveness and frequent relapse. These include the molecular hallmark translocation t(11;14)(q13;q32) CCND1/IGH, which leads to cyclin D1 overexpression and cell cycle deregulation. Other frequent genetic aberrations are mutations in crucial DNA damage response genes, such as ATM and TP53. Relevant for the present work, however, recent studies show an important role for extrinsic, soluble and adhesion-mediated signals from the tumor microenvironment in supporting MCL trafficking, homing and susceptibility to therapy.5,6
Consistent with the important role of microenvironments, primary MCL and chronic lymphocytic leukemia (CLL) cells can only be cultured in vitro for a few days before they undergo spontaneous apoptosis.7,8 If co-cultured with mesenchymal stromal cells on the other hand, the in vitro cultures can be sustained for weeks.8,9 Furthermore, stromal cells of both human and murine origin can protect MCL and CLL cells from spontaneous and drug-induced apoptosis.8,10–12 While soluble molecules secreted by stromal cells such as BAFF8,13 and CXCL1214 have been shown to increase survival in malignant B cells, the protective effect is more prominent for lymphoma cells that physically adhere to stromal cells,6,8 and direct interactions between lymphoma cells and stromal cells can induce cell cycle arrest in MCL and diffuse large B-cell lymphoma (DLBCL).15 These mechanisms, involving soluble and adhesion-mediated signaling, may specifically confer survival advantages to lymphoma cells that home to protective microenvironmental niches through the activation of anti-apoptotic programs and downregulation of genes involved in proliferation.16
Targeted cell-culture studies have elucidated effects of microenvironment interactions in MCL and CLL. Increased levels of immunomodulatory cytokines, such as CCL3, CCL4, CCL22, IL-10 and TNF, with the capacity to alter microenvironment cellular composition have been reported in co-cultures of MCL or CLL cells with stromal cells or under other conditions that mimic microenvironment interactions.17–20 The adhesive properties of non-Hodgkin lymphoma (NHL) cells have been shown to increase upon treatment with anti-IgM, CXCL12 or CXCL13.17 The CXCR4 cytokine receptor protein, central to normal B-cell migration and homing, is down-regulated in adherent CLL cells.14,21 In co-culture and analogous studies, increased expression of anti-apoptotic proteins, such as BCL-XL and MCL-1, have been reported.11,22,23 Co-cultivation of MCL cells with stromal cells has also been reported to increase protein levels of the cell cycle inhibitors p21Cip1 and p27Kip1, along with an increased ratio of G0/G1 cells relative to S-phase cells.15 Many of these effects may be associated with an adhesion-related induction of both the canonical and non-canonical NF-κB pathways.8
While important signaling mechanisms relevant for cell adhesion-mediated survival of lymphoma cells have been revealed by targeted studies, the present work is the first systematic study of global changes in gene expression in a defined model system that allows discrimination of gene expression changes in the different cell types in the co-culture as well as their relationship to the same cells grown in isolation.
Methods
Cell culture
Cells were cultivated in a humidified incubator at 37°C and 5% CO2 in media supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. The mouse stromal cell line MS-5 and the MCL cell line Jeko-1 were purchased from DSMZ and maintained in αMEM-glutamax (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (H.I. FBS; Gibco) and 2 mM sodium pyruvate or RPMI-glutamax (Gibco) supplemented with 10% HI FBS, respectively. Co-cultures of Jeko-1 with MS-5 at a 10:1 ratio were maintained under the same conditions as for MS-5 cells alone.
Cell-cell binding assay
Unlabeled Jeko-1 suspension cells were added to established MS-5 monolayers. After 24 h, unlabeled Jeko-1 cells in suspension were removed and replaced with an equivalent number of CFDA-SE labeled Jeko-1 cells. Adhered unlabeled/labeled Jeko-1 cells were counted at 24 h and 48 h. The order of addition of labeled/unlabeled Jeko-1 cells was subsequently reversed.
RNA extraction, library preparation and sequencing
Total RNA was extracted using RNeasy with QIAshredders (Qiagen). Libraries were prepared using TruSeq sample prep kit v.2.0 and included a poly-A enrichment step. Samples were 16-plexed on an Illumina HighSeq 2500 instrument generating 230,700,000 2×101bp short reads (Online Supplementary Table S1).
Species-based read separation and mapping to reference genomes
Reference genomes hg19 and mm10 were obtained from UCSC. Raw reads were separated based on species origin using Xenome (v.1.0.1).24 Separated reads of human and murine origin were aligned to reference genomes hg19 and mm10, respectively, using Tophat2 (2.0.11)25 using default options. Fragments per feature were counted using summarizeOverlaps from the Bioconductor package GenomicAlignments (1.0.6) with counting mode set to “Union”. Gene annotations and coordinates were from the Bioconductor packages TxDb.Hsapiens.UCSC.hg19.knownGene (v.2.14.0) and TxDb.Mmusculus.UCSC.mm10.knownGene (v.2.14.0).
Differential gene expression analysis and GSEA
Differential expression was determined by DESeq (1.16.0).26 Normalized count tables were used for Gene Set Enrichment Analysis (GSEA) (Broad Institute, 2.2.0)27 using canonical pathways (c2.cp.v5.0.symbols.gmt) and GO processes (c5.bp.v5.0.symbols.gmt), from the MsigDB.27 Functional clusters from the leading edge analysis were identified as ngenes ≥ 10.
Protein interaction network analysis
A unified list with unique gene identifiers based on differentially expressed genes (FDR q-value≤0.05) and leading edge genes from the GSEA (n=1458) was uploaded to the STRING interaction database (v.10.0) with the confidence level set to 0.4.28 Node genes were defined as those having 8 or more interactions.
Microarray analysis
Microarray datasets were downloaded from GEO:29 GSE2102930 and GSE70910.31 Overlaps between these datasets and adhesion-related genes from the present study were interrogated by two-sided Fisher’s exact tests.
Data availability
Raw RNA-seq data are available via the gene expression omnibus (GEO) repository29 by accession number: GSE99501. A detailed account of materials and methods used is available in the Online Supplementary Methods.
Results
MCL cells adhere stably to stromal cells
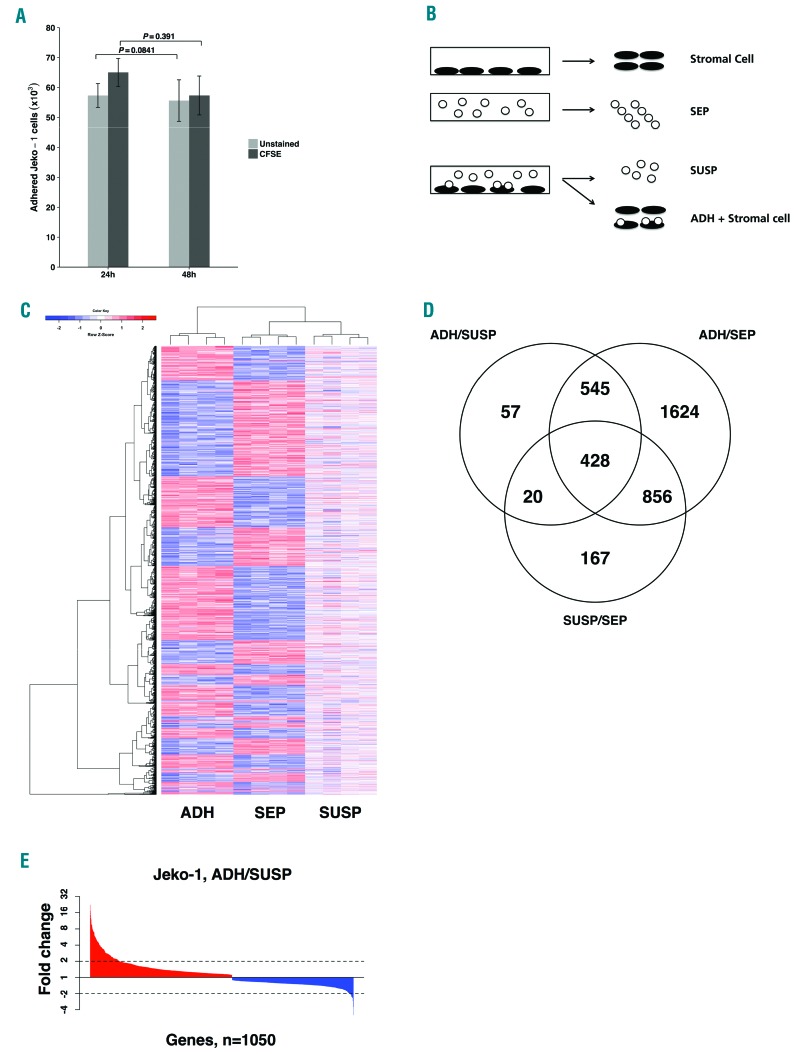
Approximately 10% of MCL cells (Jeko-1), which normally grow in suspension, adhered to a mono-layer of the bone marrow-derived, adherent stromal cell line (MS-5) upon co-culture and remained in place when MCL cells remaining in suspension were poured away after 24 h (Figure 1A). The interaction with stromal cells was stable because addition of a 10-fold excess of CFDA-SE labeled MCL cells to the stromal cells with adhered MCL cells and incubation for a further 24 h did not significantly displace the unlabeled bound cells (Figure 1A). Interestingly, the labeled MCL cells were able to bind to stromal cells independently of the previously bound MCL cells. Similar results were obtained when the order of addition of labeled and unlabeled cells was reversed (Figure 1A), indicating that the CFDA-SE label does not significantly affect the adherence characteristics of Jeko-1 cells in this assay. Thus, the co-cultured MCL cells could be divided into two relatively stable subsets (adherent and suspension), and therefore it was of interest to characterize differences in gene expression between these subsets and in relation to mono-cultured cells.
Figure 1.
Identification of differentially regulated genes in adherent co-culture cells. (A) A fraction of co-cultured Jeko-1 cells adhere to an MS-5 cell mono-layer and remain stably bound in the presence of excess suspension cells. The number of unlabeled (light gray) or CFDA-SE labeled (dark gray) Jeko-1 cells adhered to MS-5 stromal cells after 24 hours (h) was not significantly reduced after a further 24 h of incubation (48 h) with excess CFDA-SE (P=0.841) or unlabeled (P=0.391) Jeko-1 cells, respectively. Mean±Standard Deviation are shown for cultures (n=3). (B) Experimental design showing the three Jeko-1 and the two MS-5 stromal cell fractions for which RNAseq data were acquired. (C) Heatmap representation of relative transcript levels for the 3697 genes with significantly changed mRNA levels (FDR q-value ≤ 0.05) in at least one Jeko-1 cell fraction after hierarchical clustering. (D) Venn diagram illustrating number of differentially expressed genes in pair-wise comparisons between the three Jeko-1 cell fractions (FDR q-value ≤ 0.05, total number of changed genes: 3697). The three comparisons in the Venn diagram are: adherent Jeko-1 cells in co-culture (ADH) compared with suspension Jeko-1 cells in co-culture (SUSP), n=1050 (ADH/SUSP), ADH compared to mono-cultured Jeko-1 cells (SEP), n=3453 (ADH/SEP) SUSP compared to SEP n=1471 (SUSP/SEP). (E) Ranked fold changes of 1050 genes with changed transcript levels in ADH relative to SUSP Jeko-1 cells in co-culture.
Adhesion to stromal cells affects gene expression in MCL cells
Physical separation of adherent MCL cells from stromal cells in co-cultures was avoided by direct RNA isolation from the mixed cell population in order to minimize separation-associated artifacts in the RNAseq data. The mixed-species short RNAseq reads were separated in silico by species-specific read separation using reference genomes for human (MCL cells) and mouse (stromal cells). Read counts per sample prior to and following species-specific read separation are presented in Online Supplementary Table S1.
The experimental design allowed for global transcript level comparisons between three distinct MCL fractions: mono-cultured MCL cells (SEP), adherent MCL cells in co-culture (ADH), and suspension MCL cells in co-culture (SUSP) (Figure 1B). Pairwise comparisons of transcript levels for the three MCL fractions identified a total 3697 genes that are differentially expressed in at least one comparison after 24 h mono-/co-culture (FDR q-value ≤0.05). The heat map in Figure 1C shows relative transcript levels across the three cell populations. Overall, the largest difference is between the ADH fraction and the suspension cells in mono-culture (SEP) and co-culture (SUSP), which while distinct, are more similar to each other (column dendrogram, Figure 1C).
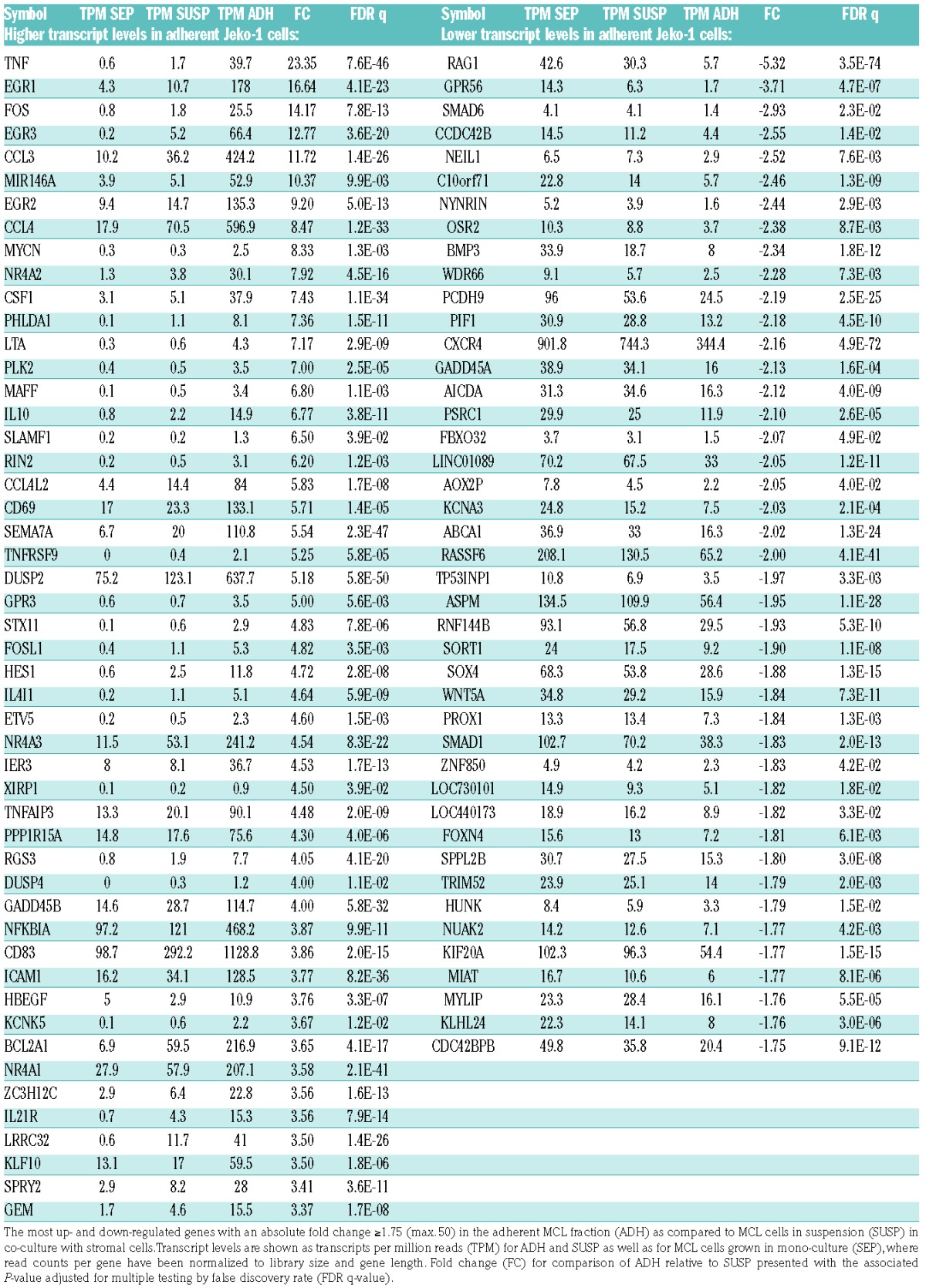
The Venn diagram sets in Figure 1D show the number of genes that are differentially expressed between each pair of MCL cell populations: 3453, 1471 and 1050 genes were differentially expressed in the respective comparisons of ADH relative to SEP (ADH/SEP), SUSP relative to SEP (SUSP/SEP) and ADH relative to SUSP (ADH/SUSP). The three comparison groups of differentially regulated genes are highly overlapping with half the genes occurring in two or more groups (Figure 1D). Given the previously reported survival advantage conferred to lymphoma cells adhering to stromal cells in co-culture,6,8 and as this comparison provides the best opportunity for specifically understanding molecular aspects associated with adhesion to stromal cells, we focused the analysis on the 1050 genes with altered transcript level between the two MCL cell fractions within the co-culture system (ADH/SUSP). Altogether, 137 of these genes were changed more than 2-fold at the transcript level (Figure 1E). Significantly changed genes in the ADH fraction with the highest fold change are presented in Table 1 (Complete lists of significantly changed genes are available in Online Supplementary Tables S2–S4).
Table 1.
Differential transcript levels in adherent compared to suspension mantle cell lymphoma (MCL) cells in co-culture.
Gene expression and co-culture dependent changes in stromal cells
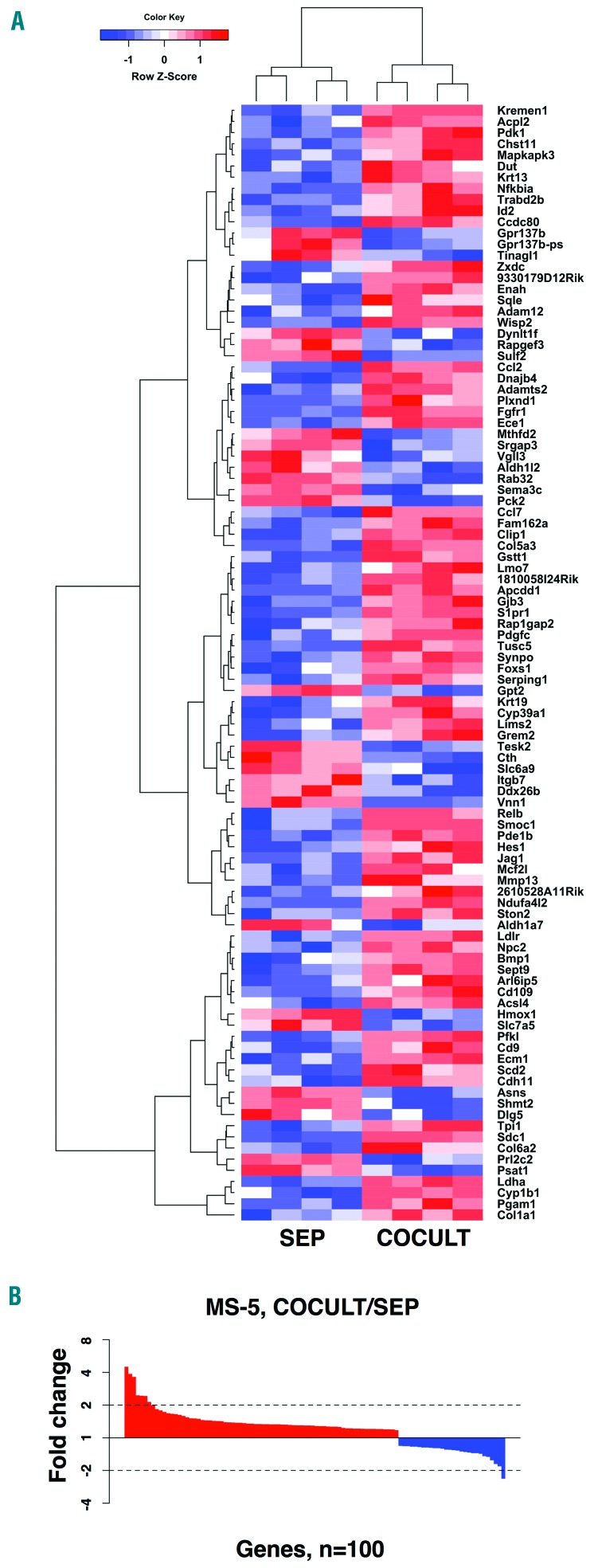
The study design did not allow for an in-depth comparison of transcript levels in stromal cells analogous to the analysis of cancer cells above. Thus a detailed analysis of differentially expressed stromal cell genes is not reported here. Nonetheless, 100 genes were significantly changed at the transcript level between mono- and co-cultured stromal cells (Figure 2). These include the chemotactic molecules Ccl2 and Ccl7, both with higher transcript levels in the co-cultured MS-5 cells. These can interact with the CCR2 cytokine receptor, which is expressed in Jeko-1 MCL cells. A complete list with significant transcript level changes between mono- and co-cultured MS-5 stromal cells is available in Online Supplementary Table S5.
Figure 2.
Significant transcript level changes between mono- and co-cultured stromal cells. (A) Heatmap representation of the 100 genes with significant transcript level changes between co-cultured (COCULT) and mono-cultured (SEP) and MS-5 stromal cells (FDR q-value ≤0.05) after hierarchical clustering. (B) Ranked fold changes for the 100 genes with altered transcript levels between COCULT and SEP MS-5 stromal cell fractions.
Adhesion dependent changes in MCL cells are associated with four main functional themes
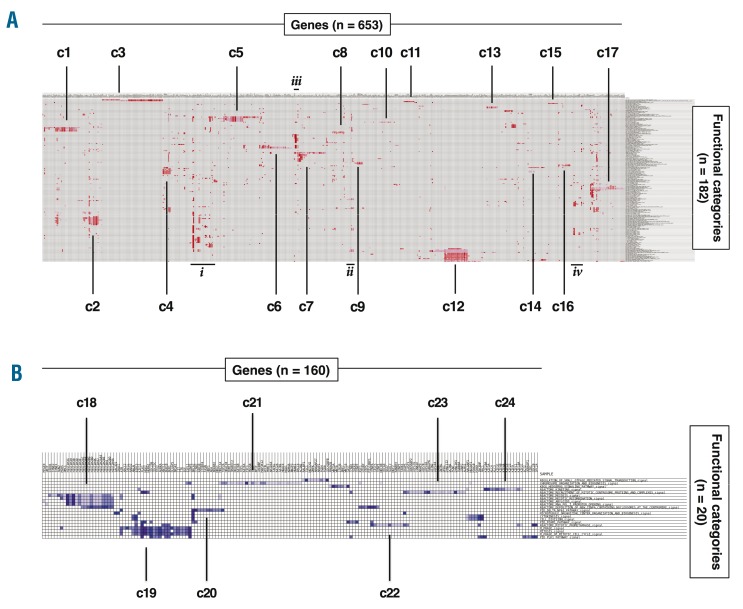
Gene set enrichment analysis was used to identify functional categories enriched in the significantly up- (182 gene sets) or down-(20 gene sets) regulated genes in MCL cells adhered to stromal cells. Leading-edge genes (n=819), which account for the level of the enrichment score for each of the functional categories, were clustered across the set of identified functional categories to produce heat maps showing clusters of genes and functional categories that together identify functional differences between adherent and suspension cells in the co-culture. Figure 3 identifies 24 functional clusters (containing at least 10 genes and ≥ 1 functional category) for genes with increased (17 clusters, Figure 3A) or decreased (7 clusters, Figure 3B) transcript levels in adherent MCL cells. The genes and functional categories defining each cluster are listed in Table 2, and full lists of leading edge genes and enriched gene sets from GSEA are presented in Online Supplementary Tables S6–S9.
Figure 3.
Identification of functionally related clusters of genes by Gene Set Enrichment Analysis leading edge analysis. Heatmaps showing clustering of functional categories and leading edge genes that are up-regulated (A) or down-regulated (B) in adherent Jeko-1 cells relative to suspension of Jeko-1 cells in co-cultures. The numbers of leading edge genes and functional categories are shown in each panel: 24 clusters (c1-c24), defined as having ≥10 genes and ≥1 functional categories, were identified (see Table 2 for more details). Four additional groups of genes that contain many functional categories but which do not fulfill the cluster criteria are denoted i–iv.
Table 2.
Functional clusters of genes distinguishing stromal-cell attached MCL cells from mantle cell lymphoma (MCL) suspension cells in co-culture.

Most functional gene clusters can be attributed to a small number of cellular themes, including B-cell activation and immune cell signaling (c2, c4, c7, c10, c17), apoptosis and anti-apoptosis (c1, c5), cell adhesion and migration (c6, c8, c15, c16), and early mitosis (c19, c22, c23, c24). The first three themes contain up-regulated genes in adherent cells while the fourth cluster contains down-regulated genes. Genes in the B-cell activation-related clusters included B-cell receptor components, co-stimulatory surface molecules and soluble factors (e.g. CD79A, CD86, CD180, ICOSLG, PDCD1 and TGFB1). The apoptosis-related clusters contained anti-apoptotic BCL2-family genes (e.g. BCL2A1, BCL2L1 and MCL1) while adhesion- and migration-related clusters included ACTB, ACTG1 and TUBA4A. The mitosis-related genes are primarily involved in early mitosis steps such as mitotic onset (e.g. AURKA, CCNB2 and PLK1), spindle establishment (e.g. BUB1, CENPA, CENPE), and kinetochore formation (e.g. KIF20A, KIF4A, KIF5A). These themes are each compatible with survival strategies resulting from adherence dependent changes in immune cell characteristics, apoptosis pathways and cell cycle status. Other clusters (c9, c11, c13, c20, c21) represent more general functional characteristics, which could in principle contribute to one or more of the process-related themes. Finally, there are 4 clusters (c3, c12, c14, c18) for which fewer than 10% of member genes are differentially regulated even though they do form part of the GSEA leading edge. These clusters may be important but they contain only 11 significantly differentially regulated genes. For example, the c12, containing proteasome subunits, is of interest given the use of the proteasome inhibitor, bortezomib, for treatment of refractory MCL.
Apart from the clustered genes there are several leading-edge genes that are not easily clustered because they occur in many functional classes (Groups i–iv, Figure 4A). Examples include NFKB1, NFKB2, ICAM1, CCL3 and CCL4 and the groups, as well as other results are detailed in the Online Supplementary Appendix. Secreted CCL3/4 levels increased in co-cultures and upon MCL-cell simulation with anti-IgM (Online Supplementary Figure S1).
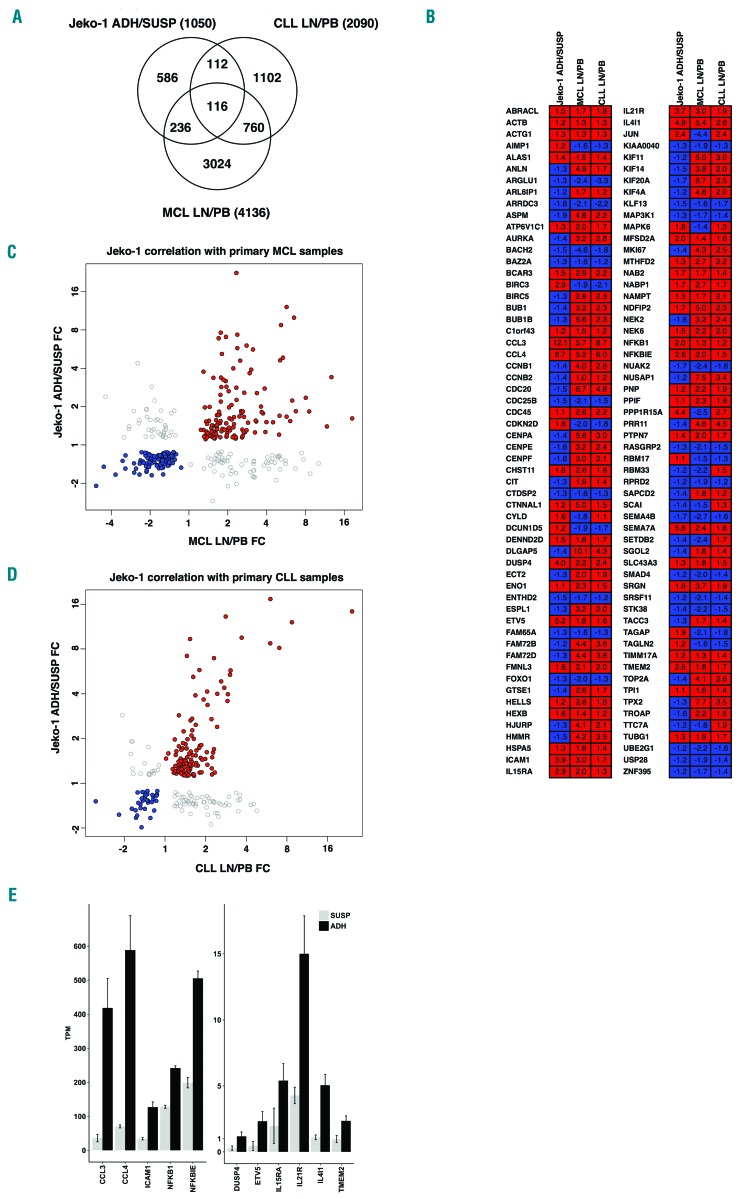
Figure 4.
Differentially expressed genes in co-culture adherent mantle cell lymphoma (MCL) cells overlap with microenvironment-regulated genes in MCL and chronic lymphocytic leukemia (CLL) patients. (A) Venn diagram showing overlaps between differentially expressed genes between adherent and suspension Jeko-1 cells in co-culture (ADH/SUSP) compared with microenvironment-regulated genes from microarray studies of MCL and CLL patients, where transcript levels in lymph nodes were compared to those of peripheral blood (data from GSE21029 and GSE70910, respectively). The numbers of differentially expressed genes (FDR q-value ≤ 0.05) for each data set are shown. (B) Comparison of the direction of regulation for the 116 genes in the overlap of all three data sets. Color-coding is red for genes up-regulated in adherent cells/lymph node and blue for down-regulated genes. Fold change values are shown for each gene and data set. (C) Scatter plot showing fold changes for 348 genes with significant transcript level changes in both adherent MCL cells relative to suspension co-culture cells (Jeko-1 ADH-SUSP FC) and MCL patient lymph node relative to peripheral blood (MCL LN-PB FC). Spearman Rho=0.843 (P=1.83×10−68) for genes with coherent directional change in the two data sets where red data points (n=135) represent up-regulated genes and blue data points (n=114) down-regulated. Gray data points (n=99) represent genes with opposite direction of regulation between the two data sets. (D) Scatter plot showing fold changes for 228 genes with significant transcript level changes in both adherent MCL cells relative to suspension co-culture cells (Jeko-1 ADH-SUSP FC) and CLL patient lymph node relative to peripheral blood (CLL LN-PB FC). Spearman Rho=0.746 (P=8.39×10−29) for genes with coherent directional change in the two data sets where red data points (n=121) represent up-regulated genes and blue data points (n=34) down-regulated. Gray data points (n=73) represent genes with opposite direction of regulation between the two data sets. (E) A core gene set of 13 cell-adhesion related microenvironmentally regulated genes in MCL and CLL (see text for details). Transcript levels of the genes in adherent (ADH, black bars) and suspension (SUSP, gray bars) MCL cells in co-cultures are plotted; mean transcripts per million reads (TPM) where the read counts are normalized to library size and feature length±Standard Deviation.
Altogether 199 of the 1050 genes with significantly altered transcript levels (19%) were functionally classified by GSEA. Network analysis connected an additional 495 adhesion regulated genes to the 24 GSEA clusters, thus functionally connecting approximately 65% of the 1050 differentially expressed genes. Network analysis did not identify any gene networks not connected to GSEA clusters, strongly suggesting that we have identified the main processes and pathways defined by differential expression of genes in adherent cells.
Overlap between adhesion-regulated MCL cell genes and microenvironment-regulated genes in MCL and CLL patients
Analysis of publicly available data from two independent studies of CLL and MCL30,31 identified 2090 and 4136 differentially expressed genes, respectively, between cells from lymph node and blood (FDR q-value≤0.05). Comparison of the 1050 adhesion-associated genes observed in the in vitro co-culture system with differentially expressed genes from the CLL and MCL datasets showed significant overlaps of 228 genes (22%, P=2.7×10−38) and 348 genes (34%, P=8.2×10−38), respectively (Figure 4A). In all, 116 genes were differentially expressed in all three datasets (Figure 4B). Forty-eight (41%) of these genes are included in the GSEA leading edge used to define cell-adhesion-related processes and 32 (28%) of these are members of one of the 24 functional clusters (Table 2), with 29 being in clusters that primarily define the four functional themes discussed previously. Thus the 116 genes that overlap between the three studies are representative of the gene clusters defining adhesion-associated gene expression. For 65 of the genes, the direction of regulation (up or down) was the same in each of the three datasets, assuming similarity between adherent cells in vitro and lymph node cells in vivo. Similarities are also indicated by fold change correlations for genes that are differentially regulated in both the in vitro co-culture system and the MCL and CLL datasets (Figure 4C and D). Although there are differences between in vivo and in vitro studies, the co-culture system faithfully reproduces a significant subset of differential gene regulation events observed in MCL and CLL patients.
Thirteen of the 65 genes with conserved direction of regulation (CCL3, CCL4, DUSP4, ETV5, ICAM1, IL15RA, IL21R, IL4I1, MFSD2A, NFKB1, NFKBIE, SEMA7A, TMEM2) had a fold-change value of 2 or more in the present study (Figure 4E). CCL3, CCL4 and NFKBIE are members of previously identified NF-κB and BCR signatures30 and are likely to represent a broader activation of NF-κB and BCR pathways (Online Supplementary Figure S2). Of the 51 genes with a different regulation direction in one of the three datasets, 43 showed a discrepancy in the in vitro data in relation to the patient studies; 38 of these genes were related to proliferation, early mitosis or mitotic spindle formation.
Discussion
Mantle cell lymphoma is a B-cell lymphoma that is difficult to cure and patients experience frequent relapses, often resulting from minimal residual disease. As described above, adherence of lymphoma cells to stromal cells within microenvironmental niches is thought to be essential for their proliferation, survival and drug resistance, as well as for their immunomodulatory ability to recruit other cell types to the microenvironment.8–11,17,18
Here, we developed a co-culture model system to systematically dissect differences in gene expression that occur in MCL cells and stromal cells that adhere to each other, using genome-wide RNA sequencing. A total of 1050 adherence-specific genes in MCL cells represent 24 functionally defined gene clusters, many of which can be attributed to four main functional themes. These correspond well with the important biological and pathological characteristics that functionally define lymphoma cells in microenvironments. The identified functions may be acquired characteristics specific to cancer cells or characteristics associated with normal B cells. The 1050 differentially regulated genes in adherent MCL cells significantly overlap with genes that are differentially expressed in lymph node, compared to blood, in MCL and CLL patients. The most differentially regulated genes included B-cell receptor signature genes that were not seen among highly regulated genes in a previous study,32 perhaps either due to the different study designs used or because different in vitro systems reflect different aspects of microenvironment interactions.
Cell adhesion, migration and homing
Mantle cell lymphoma cell adhesion to stromal cells is associated with induction of genes involved in cellular adhesion and cell motility/migration. The extracellular matrix cluster (c6) consists entirely of matrisome proteins,33 where 9 encode matrisome core proteins (glycoproteins, collagens or proteoglycans) that constitute structural components of extracellular matrix. The remaining 29 genes encode matrisome-associated proteins, which modulate extracellular matrix function, including cellular adhesion. Other clusters contain structural components important for cell rigidity and motility such as ACTB, ACTG1 and TUBA4A. Additional cell-cell adhesion-related genes with significantly higher transcript levels in adherent MCL cells include ICAM1, ITGB2 and AMIGO2. These may be important for homing of MCL cells to microenvironmental niches and their retention in the niche.
The CXCR4 gene encoding a homing-related cytokine receptor is down-regulated in adherent cells, consistent with previous reports on microenvironment-associated downregulation of its mRNA and protein level in MCL and CLL patients as well as cell culture systems.17,30,34–36
The CXCR4 ligand, CXCL12, is expressed by the stromal cells and can promote adhesion of MCL cells to fibronectin and VCAM-1.17 Thus, this receptor-ligand pair could facilitate homing and subsequent adhesion of MCL cells to microenvironments.
Anti-apoptosis and cell survival
Anti-apoptosis clusters are up-regulated in adherent cells and include genes involved in CD40 signaling (c1), which is known to have an anti-apoptotic effect on MCL cells.37 A second example (c5) includes upregulation of BCL2-family members (eg. BCL2A1, BCL2L1, and MCL1) as well as BCL2 that is up-regulated in the co-cultured MCL cells relative to mono-cultured cells. Importantly, increased level of alternative family members causes resistance to the BCL2 inhibitor ABT-737 in CLL,38 thus indicating their significance for designing therapies targeting the BCL2-family. Results expand previous knowledge showing that stromal cell interaction protects MCL and CLL cells from spontaneous and drug-induced apoptosis.8,10, 11
Cell proliferation
The observed downregulation of early mitosis genes is consistent with previous observations showing cell cycle arrest in MCL and diffuse large B-cell lymphoma (DLBCL) cells upon interaction with stromal cells in vitro15 as well as lower response rates to cytostatic drugs observed for adherent lymphoma cells compared to cells in suspension.8 Reduced proliferation of MCL cells co-cultured with stromal cells lacking CD40L relative to cells expressing CD40L37 may indicate interaction partner-specific differences in proliferative response. While adherent MCL cells up-regulate CD40, the CD40L was not detected, consistent with downregulation of cell cycle genes in this system.
Immune cell recruitment
Adherent MCL cells up-regulate genes involved in B-cell receptor (BCR) downstream signaling, seen in BCR and NF-κB gene signatures, including genes involved in immune-modulation via recruitment of immune cells, such as CCL3 and CCL4 that are known to be induced in adherent MCL cells.17,20 CCL3 and CCL4 attract activated T cells and monocytes, and CCL4 has been shown to attract regulatory T cells.39 T-cell infiltration has prognostic relevance in MCL40 and elevated serum levels of CCL3 and CCL4 are correlated with an inferior prognosis in DLBCL and CLL.41,42
A further example is the increased level of the immunoregulatory chemokine IL-10 in adherent cells, which permits autocrine survival signaling via the IL-10 receptor (IL10RA, 2.9-fold up-regulated in co-cultured cells) and STAT3 (cluster c17).43 BCR activation can induce such an autocrine survival loop in MCL which is attenuated by the proteasome-inhibitor drug, bortezomib.44 Elevated serum levels of IL-10 are associated with poor disease outcome in DLBCL.45 Regulatory B cells commonly express IL-10 and phenotypically-related CLL cells have been ascribed similar immunosuppressive attributes.46
The importance of BCR-signaling is supported by observations that blocking BCR signaling by the BTK inhibitor ibrutinib (PCI-32765) both lowered plasma levels of CCL22, CCL4, TNF and IL-10 and caused MCL cells to leave their protective microenvironmental niches and enter the peripheral blood.17 BTK-dependent secretion of the affected cytokines was induced in vitro by co-culture with stromal cells or IgM stimulation. Reduced transcript and secreted level of IL-10 has also been observed in DLBCL cell lines treated with ibrutinib or the mTOR inhibitor, AZD2014.47
Ccl2, which could play a role in monocyte recruitment and polarization similar to that shown in follicular lymphoma,48 and Ccl7, which promotes and directs the migration of macrophages,49 were up-regulated in adherent stromal cells. Thus adherence of MCL cells to stromal cells appears to be sufficient to establish the cytokine production needed for recruitment of monocytic cells and T-cell subsets even in the absence of such cells.
Utility of the co-culture model and identification of a core set of microenvironment genes
The utility of the co-culture model system is supported by the systematic identification of functional themes that correspond well with current knowledge about how lymphomas in general and MCL in particular develop and survive. Relevance was further supported by overlap of the 1050 adherence-regulated genes with microenvironment (lymph node) regulated genes in MCL and CLL patients.30,31 The overlap is similar in extent to the overlap between the MCL and CLL data, indicating that the in vitro model system reciprocates relevant aspects of microenvironment-mediated gene regulation in lymphoma cells. The direction of regulation (up or down) was not conserved between the three datasets in 51 of the 116 genes that overlap between all three datasets, and, perhaps unsurprisingly, it is most often the in vitro data (43 of 116 genes) that differ from the in vivo studies. Interestingly, most of these genes are associated with the cell cycle, consistent with previous reports of cell cycle arrest in MCL and DLBCL cells adhering to stromal cells.15 The effect of cell adherence on the cell cycle may depend on specific properties of the interacting stromal cells, as discussed above in relation to expression of CD40L.37
Among 116 overlapping genes, a core gene set of 13 genes (fold change >2-fold) was identified. The core gene set contains genes that relate to the four functional themes and that have previously been coupled to lymphoma pathogenesis (CCL3, CCL4, ICAM1, NFKB1 and IL21R) as well as DUSP4, ETV5, IL15RA, IL4I1 and NFKBIE that have been described in a lymphoma context. The presence of the NFKBIE NF-κB inhibitor may appear contradictory, but the apoptotic/anti-apoptotic responses to the NF-κB pathway activity have been shown to be pluralistic and context dependent.50 The set also includes three genes which have not previously been associated with lymphoma pathology, MFSD2A, SEMA7A and TMEM2, suggesting the existence of at least some relevant genes that remain to be characterized.
In summary, this first genome-wide, systematic study shows that genes differentially expressed in stromal cell adherent MCL cells are predominantly involved in anti-apoptosis, B-cell signaling, cell adhesion and early mitosis. Overlaps with clinical MCL and CLL data sets suggest that the identified genes also play important roles within cancer microenvironments in patients. The results support the utility of this in vitro model system for dissecting microenvironmental signaling and identify a list of 13 critical genes that should be a focus for future studies.
Supplementary Material
Acknowledgments
We would like to thank the core facility at Novum, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska hospital.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/4/666
Funding
The study was supported by grants from: The Swedish Research Council (to AW), The Swedish Cancer Society (to AW and BS), The Cancer Society in Stockholm (to BS), The Stockholm County Council (to BS) and Karolinska Institutet (to AW and BS).
References
- 1.Perez-Galan P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117(1):26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow S, Campo E, Harris N, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Vol. 2 IARC; 2008. [Google Scholar]
- 3.Herrmann A, Hoster E, Zwingers T, et al. Improvement of overall survival in advanced stage mantle cell lymphoma. J Clin Oncol. 2009;27(4):511–518. [DOI] [PubMed] [Google Scholar]
- 4.Geisler CH, Kolstad A, Laurell A, et al. Long-term progression-free survival of mantle cell lymphoma after intensive front-line immunochemotherapy with in vivo-purged stem cell rescue: a nonrandomized phase 2 multicenter study by the Nordic Lymphoma Group. Blood. 2008;112(7):2687–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23(1):43–52. [DOI] [PubMed] [Google Scholar]
- 6.Kurtova AV, Tamayo AT, Ford RJ, Burger JA. Mantle cell lymphoma cells express high levels of CXCR4, CXCR5, and VLA-4 (CD49d): importance for interactions with the stromal microenvironment and specific targeting. Blood. 2009;113(19):4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins RJ, Verschuer LA, Harmon BV, et al. Spontaneous Programmed Death (Apoptosis) of B-Chronic Lymphocytic-Leukemia Cells Following Their Culture Invitro. Br J Haematol. 1989;71(3):343–350. [DOI] [PubMed] [Google Scholar]
- 8.Medina DJ, Goodell L, Glod J, et al. Mesenchymal stromal cells protect mantle cell lymphoma cells from spontaneous and drug-induced apoptosis through secretion of B-cell activating factor and activation of the canonical and non-canonical nuclear factor kappaB pathways. Haematologica. 2012;97(8):1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand AV. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocyt ic leukaemia cells in vitro. Br J Haematol. 1996;92(1):97–103. [DOI] [PubMed] [Google Scholar]
- 10.Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998;91(7):2387–2396. [PubMed] [Google Scholar]
- 11.Kurtova AV, Balakrishnan K, Chen R, et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood. 2009;114(20):4441–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weekes CD, Pirruccello SJ, Vose JM, Kuszynski C, Sharp JG. Lymphoma cells associated with bone marrow stromal cells in culture exhibit altered growth and survival. Leuk Lymphoma. 1998;31(1–2):151–165. [DOI] [PubMed] [Google Scholar]
- 13.Lwin T, Crespo LA, Wu A, et al. Lymphoma cell adhesion-induced expression of B cell-activating factor of the TNF family in bone marrow stromal cells protects non-Hodgkin’s B lymphoma cells from apoptosis. Leukemia. 2009;23(1):170–177. [DOI] [PubMed] [Google Scholar]
- 14.Burger JA, Tsukada N, Burger M, et al. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96(8):2655–2663. [PubMed] [Google Scholar]
- 15.Lwin T, Hazlehurst LA, Dessureault S, et al. Cell adhesion induces p27Kip1-associated cell-cycle arrest through down-regulation of the SCFSkp2 ubiquitin ligase pathway in mantle-cell and other non-Hodgkin B-cell lymphomas. Blood. 2007;110(5):1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517–534. [DOI] [PubMed] [Google Scholar]
- 17.Chang BY, Francesco M, De Rooij MF, et al. Egress of CD19(+)CD5(+) cells into peripheral blood following treatment with the Bruton tyrosine kinase inhibitor ibrutinib in mantle cell lymphoma patients. Blood. 2013;122(14):2412–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard S, Danglade D, Gardano L, et al. Inhibitors of BCR signalling interrupt the survival signal mediated by the microenvironment in mantle cell lymphoma. Int J Cancer. 2015;136(12):2761–2774. [DOI] [PubMed] [Google Scholar]
- 19.Schulz A, Toedt G, Zenz T, et al. Inflammatory cytokines and signaling pathways are associated with survival of primary chronic lymphocytic leukemia cells in vitro: a dominant role of CCL2. Haematologica. 2011;96(3):408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger JA, Quiroga MP, Hartmann E, et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113(13):3050–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94(11):3658–3667. [PubMed] [Google Scholar]
- 22.Chiron D, Dousset C, Brosseau C, et al. Biological rational for sequential targeting of Bruton tyrosine kinase and Bcl-2 to overcome CD40-induced ABT-199 resistance in mantle cell lymphoma. Oncotarget. 2015;6(11):8750–8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen IM, Kitada S, Leoni LM, et al. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 2002;100(5):1795–1801. [PubMed] [Google Scholar]
- 24.Conway T, Wazny J, Bromage A, et al. Xenome–a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28(12):i172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herishanu Y, Perez-Galan P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117(2):563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saba NS, Liu D, Herman SE, et al. Pathogenic role of B-cell receptor signaling and canonical NF-kappaB activation in mantle cell lymphoma. Blood. 2016;128(1):82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivina M, Hartmann E, Vasyutina E, et al. Stromal cells modulate TCL1 expression, interacting AP-1 components and TCL1-targeting micro-RNAs in chronic lymphocytic leukemia. Leukemia. 2012;26(8):1812–1820. [DOI] [PubMed] [Google Scholar]
- 33.Naba A, Clauser KR, Hoersch S, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11(4):M111 014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittal AK, Chaturvedi NK, Rai KJ, et al. Chronic lymphocytic leukemia cells in a lymph node microenvironment depict molecular signature associated with an aggressive disease. Mol Med. 2014;20(290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghobrial IM, Bone ND, Stenson MJ, et al. Expression of the chemokine receptors CXCR4 and CCR7 and disease progression in B-cell chronic lymphocytic leukemia/small lymphocytic lymphoma. Mayo Clin Proc. 2004;79(3):318–325. [DOI] [PubMed] [Google Scholar]
- 36.Purroy N, Abrisqueta P, Carabia J, et al. Co-culture of primary CLL cells with bone marrow mesenchymal cells, CD40 ligand and CpG ODN promotes proliferation of chemoresistant CLL cells phenotypically comparable to those proliferating in vivo. Oncotarget. 2015;6(10):7632–7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiron D, Bellanger C, Papin A, et al. Rational targeted therapies to overcome microenvironment-dependent expansion of mantle cell lymphoma. Blood. 2016;128(24):2808–2818. [DOI] [PubMed] [Google Scholar]
- 38.Vogler M, Butterworth M, Majid A, et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood. 2009;113(18):4403–4413. [DOI] [PubMed] [Google Scholar]
- 39.Bystry RS, Aluvihare V, Welch KA, Kallikourdis M, Betz AG. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol. 2001;2(12):1126–1132. [DOI] [PubMed] [Google Scholar]
- 40.Nygren L, Wasik AM, Baumgartner-Wennerholm S, et al. T-cell levels are prognostic in mantle cell lymphoma. Clin Cancer Res. 2014;20(23):6096–6104. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi K, Sivina M, Oki Y, et al. Serum CCL3 and CCL4 Levels Function As Novel Prognostic Markers in Diffuse Large B Cell Lymphoma. Blood. 2012;120(21):2.22767573 [Google Scholar]
- 42.Sivina M, Hartmann E, Kipps TJ, et al. CCL3 (MIP-1alpha) plasma levels and the risk for disease progression in chronic lymphocytic leukemia. Blood. 2011;117(5):1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beguelin W, Sawh S, Chambwe N, et al. IL10 receptor is a novel therapeutic target in DLBCLs. Leukemia. 2015;29(8):1684–1694. [DOI] [PubMed] [Google Scholar]
- 44.Baran-Marszak F, Boukhiar M, Harel S, et al. Constitutive and B-cell receptor-induced activation of STAT3 are important signaling pathways targeted by bortezomib in leukemic mantle cell lymphoma. Haematologica. 2010;95(11):1865–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lech-Maranda E, Bienvenu J, Broussais-Guillaumot F, et al. Plasma TNF-alpha and IL-10 Level-Based Prognostic Model Predicts Outcome of Patients with Diffuse Large B-Cell Lymphoma in Different Risk Groups Defined by the International Prognostic Index. Arch Immunol Ther Exp. 2010;58(2):131–141. [DOI] [PubMed] [Google Scholar]
- 46.DiLillo DJ, Weinberg JB, Yoshizaki A, et al. Chronic lymphocytic leukemia and regulatory B cells share IL-10 competence and immunosuppressive function. Leukemia. 2013;27(1):170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ezell SA, Mayo M, Bihani T, et al. Synergistic induction of apoptosis by combination of BTK and dual mTORC1/2 inhibitors in diffuse large B cell lymphoma. Oncotarget. 2014;5(13):4990–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guilloton F, Caron G, Menard C, et al. Mesenchymal stromal cells orchestrate follicular lymphoma cell niche through the CCL2-dependent recruitment and polarization of monocytes. Blood. 2012;119(11):2556–2567. [DOI] [PubMed] [Google Scholar]
- 49.Xuan W, Qu Q, Zheng B, Xiong S, Fan GH. The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J Leukoc Biol. 2015;97(1):61–69. [DOI] [PubMed] [Google Scholar]
- 50.Staudt LM. Oncogenic Activation of NF-kappa B. Cold Spring Harbor Perspect Biol. 2010;2(6):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq data are available via the gene expression omnibus (GEO) repository29 by accession number: GSE99501. A detailed account of materials and methods used is available in the Online Supplementary Methods.