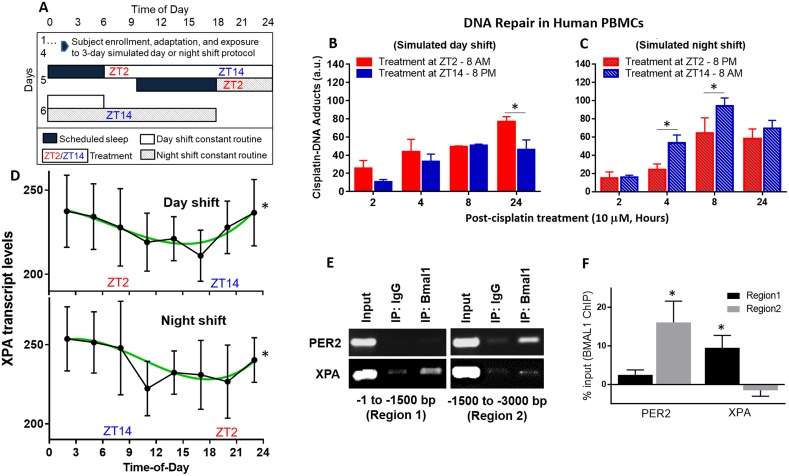
Figure 4. Repair of cisplatin-DNA adducts by time of day in human blood samples.
(A) Healthy human subjects were studied in-laboratory and subjected to 3 days of a simulated day shift schedule (B, control condition) or a simulated night shift condition (C, experimental condition). This was followed by a 24-hour constant routine protocol during which blood was drawn at 3-hour intervals (ZT2, ZT5, etc.) Blood samples collected at ZT2 and ZT14 (8 AM and 8 PM, respectively, in the day shift condition, or 8 PM and 8 AM in the night shift condition) were immediately treated with 10 μM cisplatin. Blood samples were incubated and fractions were collected between 2 and 24 hours later to isolate PBMCs. Genomic DNA was purified and probed for cisplatin-DNA adduct levels with an α-Pt-(GpG) antibody using a slot-blot assay. (D) mRNA was isolated from the blood samples and gene expression for XPA, the rate-limiting factor in NER, was analyzed using the NanoString multiplex assay. (E) DNA-protein interaction between the Bmal1 and XPA is shown in the first 3,000 base pair promoter region of human melanoma SKMEL-27 cells using a ChIP assay. PER2 is a circadian clock positive control. Input and IgG are experimental positive and negative controls, respectively. (F) Quantitation of Bmal1 binding to promoter regions of PER2 and XPA from ChIP assay, indicating regions of significance after IgG binding subtraction. Statistical analysis was done using two-way ANOVA with n=3 subjects per group (B-C) and cosinor analysis with n=7 subjects per group (D), and t test with n=3 replicates for E-F. *=p<0.05 for circadian rhythmicity or ChIP binding. Error bars = S.E.M.