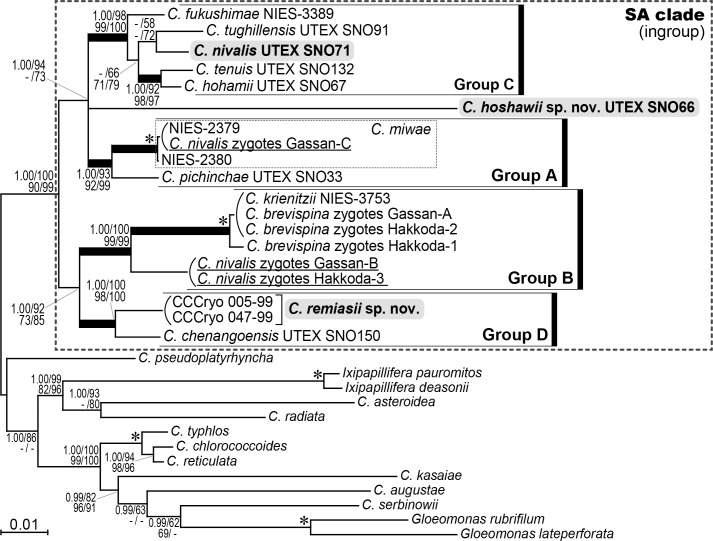
Fig 4. Bayesian phylogenetic tree of snow-inhabiting Chloromonas spp. based on 5,497 base pairs from 18S and 26S rDNA, and the first and the second codon positions of atpB and psaB.
C. nivalis zygote specimens (field-collected samples) are underlined. Corresponding posterior probabilities (PP; 0.95 or more) are shown at top left. Numbers shown in top right, bottom left and bottom right indicate bootstrap values (BV; 50% or more) in maximum likelihood (ML), maximum parsimony (MP) and neighbor-joining (NJ) analyses, respectively. Branches within the SA clade (recovered at 1.00 PP and 90% or more BV in ML, MP and NJ analyses) are shown by thick lines. Asterisk indicates 1.00 PP in BI and 100% BV in ML, MP and NJ analyses.