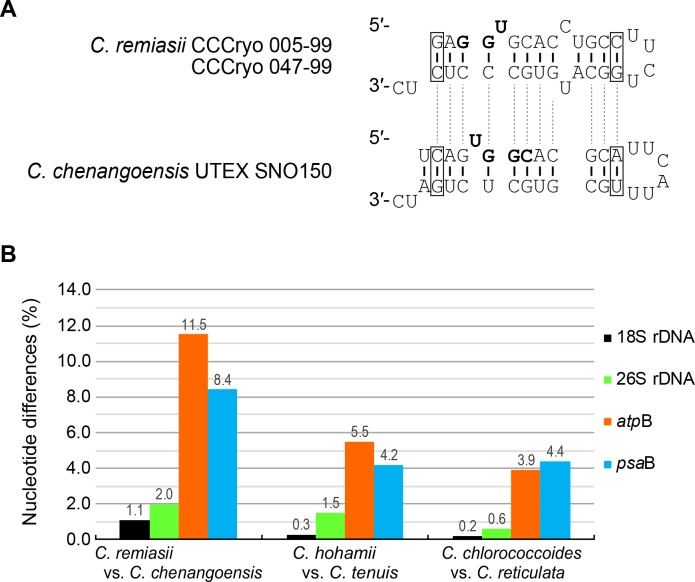
Fig 5. Genetic differences between Chloromonas remiasii Matsuzaki et al. sp. nov. and C. chenangoensis Hoham et al.
(A) Comparison of the most conserved region (near the apex of helix III encompassing the YGGY motif) of nuclear rDNA ITS2 secondary structures. Open box indicates compensatory base change. Boldface marks the YGGY motif. For the complete nuclear rDNA ITS2 secondary structures, see S6 and S7 Figs. (B) Nucleotide differences (%) from pairwise comparisons in four genes. Black: nuclear-encoded 1,748 bases of 18S ribosomal DNA (rDNA). Green: nuclear-encoded 2,020 bases of 26S rDNA. Red: chloroplast-encoded 1,128 bases of ATP synthase beta subunit gene (atpB). Blue: chloroplast-encoded 1,392 bases of P700 chlorophyll a apoprotein A2 gene (psaB). Note that the sequences from Chloromonas remiasii strains CCCryo 005–99 and CCCryo 047–99 were identical. The nucleotide differences between snow-inhabiting and mesophilic sister species [C. hohamii H.U. Ling et Seppelt vs. C. tenuis Matsuzaki et Nozaki; and C. chlorococcoides (H. Ettl et K. Schwarz) Matsuzaki et al. vs. C. reticulata (Goroschankin) Gobi] are according to the previous study [5].