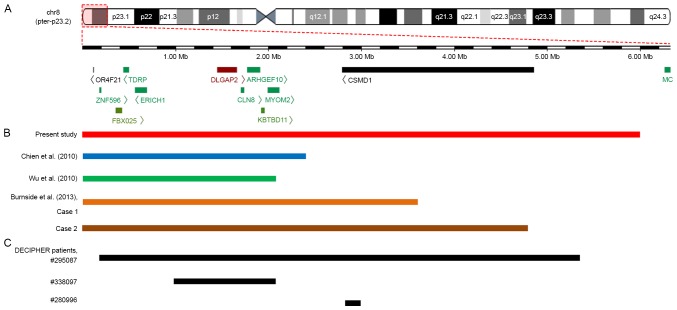
Figure 3.
An overlapping map of 8p23.2-pter deletions in the CMA-identified patients reported in the present study, previous studies and the DECIPHER database. (A) Schematic presentation of the 8p23.2-pter region and the genes located in this region, as determined by the University of California Santa Cruz Genome Browser. (B) An overview of CMA-identified patients with a deletion involving 8p23.2 to 8pter who were described in various reports. Among five patients, the deleted segment in the patient reported by Wu et al (19) was the smallest. Both deletions reported by Chien et al (17) and the deletion reported by Wu et al (19) encompassed DLGAP2, CLN8 and ARHGEF10 genes, whereas both deletions reported by Burnside et al (7) and the current study not only covered the DLGAP2, CLN8 and ARHGEF10 genes but also the CSMD1 gene. (C) An overview of patients from the DECIPHER database with deletions involving the 8p23.2 to the 8pter region that were regarded as definitely or likely pathogenic. A total of three patients presented with ID, and patient no. 338097 also exhibited behavioural problems (not specifically described). The deleted region in patient no. 338097 covered the DLGAP2, CLN8 and ARHGEF10 genes, whereas the deletion in patient no. 280996 only partially overlapped with the CSMD1 gene. CMA, chromosomal microarray analysis.