Figure 3. The transcription of rDNA starts prior to the nucleolus formation in the Drosophila embryos.
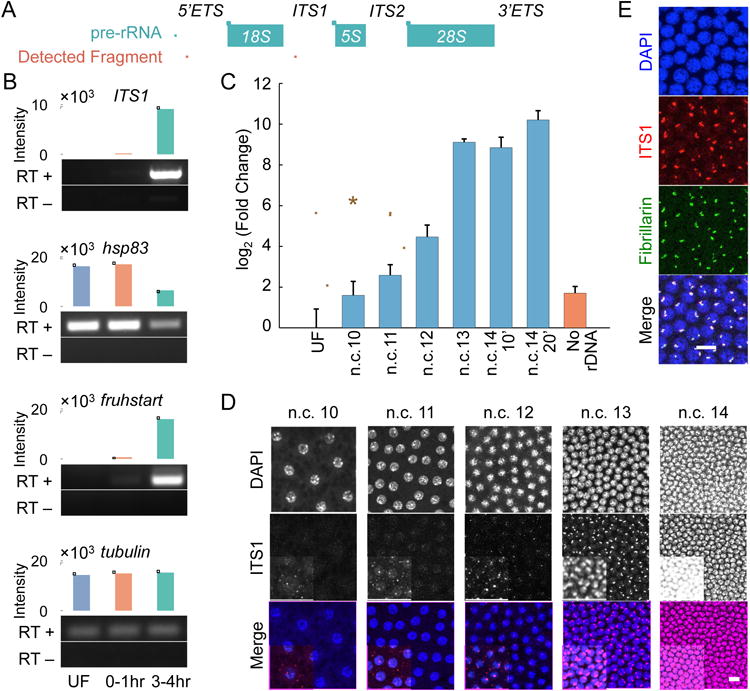
A. A schematic view of pre-rRNA, and the fragments used for detecting pre-rRNA. B. The pools of RNA present in wild-type embryos from three different collections were tested for the presence of pre-rRNA (ITS1), and three controls. The unfertilized embryos (UF) have only maternal RNA, the 0-1 hr collection has largely maternal or early zygotic RNA, and the 3-4 hr collection has mostly zygotically transcribed RNA. RT+ represents the samples that were reverse transcribed, while RT- is the negative control. hsp83 and fruhstart are maternal and zygotic, respectively. Tubulin is the loading control. C. The real-time PCR quantification of cDNAs from single embryo RNA extracts at different time points tested for the presence of 5′ETS shows a significant increase compared to the unfertilized embryos starting from n.c. 11. Data are presented as mean ± SEM of triplicate samples. Embryos were collected at seven minutes after the telophases of n.c. 10-12, 10 minutes of n.c. 13, and 10 and 20 minutes of n.c. 14. The mutants lacking rDNA were collected at n.c. 14. D. Likewise, FISH of ITS1 at n.c. 10-14 shows the emergence of bright foci starting from n.c. 11. For the purpose of presentation, the images in D and E have adjusted contrasts and changed saturation values, with the insets having lower saturation values. E. Immunolabeling of fibrillarin followed by FISH of ITS1 in embryos at n.c. 13. Scale bars 10 μm.
